For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPKSAKRRLITAFMAAGLVGGLVMSPSASADPEVPAPPVPVEPAAPPAPPNPFAFPPPAAPAAVGPADPA TPGATATDPDAQVVTNYGAPAVPIEGVPEGQNPEPFTGQPPFLPPSFNPVNGSMVGVAKPIYINFQRPIA NKQMAQDAIRITSNPPVPGRFYWVTDTQLRWRPQDFWPSNTVVNIDAAGTKSSFRVGDYLVATVDDSTHQ MEIRRNGKLEKTFPVSMGKNDGKHETKNGTYYVLEKFEDIVMDSSTYGVPVDSAEGYKLKVKDAVRIDNS GIFVHSAPWSVGDQGKRNVSHGCINLSPENARWFYEQFGSGDPVVIKNSKGGWYDQPDGASDWQMF
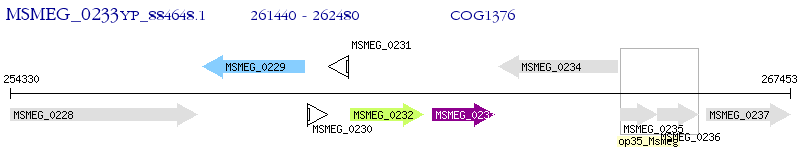
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0233 | - | - | 100% (346) | lipoprotein Lpps |
| M. smegmatis MC2 155 | MSMEG_4745 | - | 4e-60 | 43.67% (245) | ErfK/YbiS/YcfS/YnhG family protein |
| M. smegmatis MC2 155 | MSMEG_3528 | - | 6e-51 | 41.63% (209) | ErfK/YbiS/YcfS/YnhG family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0198 | - | 1e-115 | 61.10% (347) | hypothetical protein Mb0198 |
| M. gilvum PYR-GCK | Mflv_0479 | - | 1e-128 | 67.05% (349) | ErfK/YbiS/YcfS/YnhG family protein |
| M. tuberculosis H37Rv | Rv0192 | - | 1e-114 | 64.42% (312) | hypothetical protein Rv0192 |
| M. leprae Br4923 | MLBr_00426 | lppS | 4e-61 | 42.44% (271) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_4537c | - | 4e-98 | 52.91% (344) | hypothetical protein MAB_4537c |
| M. marinum M | MMAR_0435 | - | 1e-116 | 62.14% (346) | secretory protein |
| M. avium 104 | MAV_4986 | - | 1e-104 | 56.29% (318) | ErfK/YbiS/YcfS/YnhG family protein |
| M. thermoresistible (build 8) | TH_3385 | - | 1e-118 | 68.97% (290) | PUTATIVE ErfK/YbiS/YcfS/YnhG family protein |
| M. ulcerans Agy99 | MUL_1085 | - | 1e-115 | 61.76% (340) | secretory protein |
| M. vanbaalenii PYR-1 | Mvan_0177 | - | 1e-135 | 69.71% (350) | ErfK/YbiS/YcfS/YnhG family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0198|M.bovis_AF2122/97 --------MSRWKQGWTRG-SLFAALNIAAVVAV---LMLGAGVAVADPD
Rv0192|M.tuberculosis_H37Rv --------MPHWAEERHRRESNYVALEAGLDEGE---SIRRSEHSRSGCG
MMAR_0435|M.marinum_M --------MSRWTRGRIRG-SLFAAVNAAGVVGV---LLLGAGPAVADPD
MUL_1085|M.ulcerans_Agy99 --------MSRWMRGRIRG-SLFAAVNAAGVVGV---LLLGAGPAVADPD
MAV_4986|M.avium_104 --------MSGWTRG-----TLFAALNAAVVSVVGLALVLSAGPALADPD
Mflv_0479|M.gilvum_PYR-GCK --------MRQRNRKRVMALLMTAGVVAGLTLG----------APSALAQ
Mvan_0177|M.vanbaalenii_PYR-1 --------MRQRNRGRLIALFMAVGVVAGLTLG----------APSALAQ
TH_3385|M.thermoresistible__bu -----------------------------LPPD----------HPAAVSA
MSMEG_0233|M.smegmatis_MC2_155 --------MPKSAKRRLITAFMAAGLVGGLVMSP-----SASADPEVPAP
MAB_4537c|M.abscessus_ATCC_199 -----MLVLTRWFRRALVSGVCVAGLLGAGVVP-----------ALADPG
MLBr_00426|M.leprae_Br4923 MGTATRRVQPKAWRALLTLLVISVVMPGVACNRGGGNVPVNVIGDKGTPF
Mb0198|M.bovis_AF2122/97 AAPGDP---GGPG----------GPGGTAGPVDPPAVDLLAPPPDPLALP
Rv0192|M.tuberculosis_H37Rv ADAGCWRCRGGPGRGSRRSRRSRGPGGTAGPVDPPAVDLLAPPPDPLALP
MMAR_0435|M.marinum_M DAPGDP--------------------PVIAPAEP-APDPLAPPPGPLALP
MUL_1085|M.ulcerans_Agy99 DAPGDP--------------------PVIAPAEP-APDPLAPPPGPLALP
MAV_4986|M.avium_104 PAPADP-------------------GAVAAPPGP------PAPPDPLAPP
Mflv_0479|M.gilvum_PYR-GCK PAPAPA---------------------PAPVPPP----------------
Mvan_0177|M.vanbaalenii_PYR-1 PAPAPA---------------------PAPPPPPPANPFVFPNPLAPPAP
TH_3385|M.thermoresistible__bu PAPFNP---------------------FAPQP------------------
MSMEG_0233|M.smegmatis_MC2_155 PVPVEP---------------------AAPPAPPNPFAFPPPAAPAAVGP
MAB_4537c|M.abscessus_ATCC_199 DPPDPG---------------------IEEPGPP------PPPPLPWELP
MLBr_00426|M.leprae_Br4923 ADLLVPKLTASVTDGAVGVNVDMPVTVTVADGVLAAVTMINDNGRMINGQ
Mb0198|M.bovis_AF2122/97 PALDPLAPPP-PDPLAPPPPDPLAVPVAAGPVAGQDPTPFVGPPP-FRPP
Rv0192|M.tuberculosis_H37Rv PALDPLAPPP-PDPLAPPPPDPLAVPVAAGPVAGQDPTSFVGPPP-FRPP
MMAR_0435|M.marinum_M PMPDPLAPPPFPDPLAPP----PLVPVAAGPVAGQDPTPFFGPPP-FRPP
MUL_1085|M.ulcerans_Agy99 PMPDPLAPP-------------PLVPVAAGPVAGQDPTPFFGPPP-FRPP
MAV_4986|M.avium_104 PPPDPLAPPPPAAPPAPWLP--PAAQPAAAPAAGQDPTPFTGTPP-FGPP
Mflv_0479|M.gilvum_PYR-GCK --ATATGARTPATADPFAVIPGTPQPIPEGTPAGQNPNPFVGQPP-FVPP
Mvan_0177|M.vanbaalenii_PYR-1 PAPAPAGAAAPAAANPLAVIPGNPLPIPEGTPAGQNPNPFVGQPP-FIPP
TH_3385|M.thermoresistible__bu -------------------------TIPPDTPAGQNPLPYTGEPV-FLPP
MSMEG_0233|M.smegmatis_MC2_155 ADPATPGATATDPDAQVVTNYGAPAVPIEGVPEGQNPEPFTGQPP-FLPP
MAB_4537c|M.abscessus_ATCC_199 PPPPPPAPDAPPPAEVPP----VPAPVPTTPPPGAPPTGDVPPLP-VGLL
MLBr_00426|M.leprae_Br4923 FSPDGLRWSTTEPLGFNRCYTLSAKALGLGGVVNRQMTFQTSSPAHLTMP
. .
Mb0198|M.bovis_AF2122/97 TFNPVDGAMVGVAKPIVINFAVPIADRAMAESAIHISSIPPVPGKFYWMS
Rv0192|M.tuberculosis_H37Rv TFNPVDGAMVGVAKPIVINFAVPIADRAMAESAIHISSIPPVPGKFYWMS
MMAR_0435|M.marinum_M SFNPVDGAMVGVAKPIIINFQVPIADQAMAESAIHISSIPPVPGKFYWMT
MUL_1085|M.ulcerans_Agy99 SFNPVDGAMVGVAKPIIINFQVPIADQAMAESAIHISSIPPVPGKFYWMT
MAV_4986|M.avium_104 TFVPKTGSTVGVAQPIIINFPGRVDDAGAAISAVHVSSVPPVPGKFYWMT
Mflv_0479|M.gilvum_PYR-GCK SFNPVNGSIAGAAKPIYINFQRPIANRQMAQDAVHISSNPPVPGRFYWVT
Mvan_0177|M.vanbaalenii_PYR-1 SFNPSNGSIVGAAKPIYINFQRPIANRQMAQDAVHISSNPPVPGRFYWVN
TH_3385|M.thermoresistible__bu SFNPVNGALVGVAKPISIKFQRPIANRKMAEDAIHISSEPPVPGRFYWIS
MSMEG_0233|M.smegmatis_MC2_155 SFNPVNGSMVGVAKPIYINFQRPIANKQMAQDAIRITSNPPVPGRFYWVT
MAB_4537c|M.abscessus_ATCC_199 KNSPNNGDVVGVAQPITIVFAAPVTDKKAAEAAVKITTSKPAPGYFYWYT
MLBr_00426|M.leprae_Br4923 YVNPGNGEIVGIGEPVAIRFDENIANRLAAQKAITITANPPVEGAFYWLN
* * .* .:*: * * : : * *: ::: *. * *** .
Mb0198|M.bovis_AF2122/97 PTQVRWRPFEFWPANTAVNIDAAG--------------TKSSFRTGDSLV
Rv0192|M.tuberculosis_H37Rv PTQVRWRPFEFWPANTAVNIDAAG--------------TKSSFRTGDSLV
MMAR_0435|M.marinum_M PTQVRWRPFQFWPANTAVNIDAAG--------------TKSSFRTGDSLV
MUL_1085|M.ulcerans_Agy99 PTQVRWRPFQFWPANTAVNIDAAG--------------TKSSFRTGDSLV
MAV_4986|M.avium_104 PTQLRWRPLSFWPAHTAVTVDAGG--------------TVTNFQTGDTLV
Mflv_0479|M.gilvum_PYR-GCK DTQLRWRPQDFWPAGTVVTIDAAG--------------TKSSFTVPEQLT
Mvan_0177|M.vanbaalenii_PYR-1 DTQLRWRPQDFWPAGTVVTIDAAG--------------TKSTFSVPEQLV
TH_3385|M.thermoresistible__bu DTEVRWRPQDFWPAHTVVNIDAGG--------------TKSSFRVGESQV
MSMEG_0233|M.smegmatis_MC2_155 DTQLRWRPQDFWPSNTVVNIDAAG--------------TKSSFRVGDYLV
MAB_4537c|M.abscessus_ATCC_199 DQQLRWKPTQFWPANTDVNVNAGG--------------TKWSFKVGDAFV
MLBr_00426|M.leprae_Br4923 NREVRWRPEHFWKSGTAVDVAVNTYGVDLGEGMFGEDNVKTHFTIGDEVF
::**:* ** : * * : . . * :
Mb0198|M.bovis_AF2122/97 ATADDATHQMTITRNGVVQKTFPMSMGMVSGGHQTPNGTYYVLEKFATVV
Rv0192|M.tuberculosis_H37Rv ATADDATHQMTITRNGVVQKTFPMSMGMVSGGHQTPNGTYYVLEKFATVV
MMAR_0435|M.marinum_M ATADDATHQMTITRNGVVEQTFPMSMGMAAGNHQTPNGTYYVLEKMPTVV
MUL_1085|M.ulcerans_Agy99 ATADDATHQMTITRNGVVEQTFPMSMGMAAGNHQTPNGTYYVLEKMPTVV
MAV_4986|M.avium_104 ATADDATHQLTVTRNGTVEKTFPMSMGMTAGNHQTPNGTYYVQDKKASVV
Mflv_0479|M.gilvum_PYR-GCK ATIDNDTKQMEVHRNGVLEKTFPVSMGKPG--YDTKNGTYYVLEKFDEIV
Mvan_0177|M.vanbaalenii_PYR-1 ATIDNDTKQMEVVRNGVVEKTFPVSMGKSG--YDTKNGTYYVLEKFADIV
TH_3385|M.thermoresistible__bu ATIDDKTKQMEVVINGELVKTMPVSLGKKG--YETKNGTYYVLEKFKDIV
MSMEG_0233|M.smegmatis_MC2_155 ATVDDSTHQMEIRRNGKLEKTFPVSMGKNDGKHETKNGTYYVLEKFEDIV
MAB_4537c|M.abscessus_ATCC_199 STVDDATYTMTVTRNGVVERTMPISMGKHDKKHETKNGTYYVSEKFQKMV
MLBr_00426|M.leprae_Br4923 ATADDATKMLTVRVNGEVVKIMPTSMGKDS--TPTANGIYIVGARFKHII
:* *: * : : ** : : :* *:* * ** * * : ::
Mb0198|M.bovis_AF2122/97 MDSSTYGVPVNSA--QGYKLTVSDAVRIDNSGNFVHSAPWSVADQGKRNV
Rv0192|M.tuberculosis_H37Rv MDSSTYGVPVNSA--QGYKLTVSDAVRIDNSGNFVHSAPWSVADQGKRNV
MMAR_0435|M.marinum_M MDSSTYGVPVNSP--QGYKVTVADAVRIDNSGNFVHSAPWSVGDQGKRDV
MUL_1085|M.ulcerans_Agy99 MDSSTYGVPVNSP--QGYKVTVADAVRIDNSGNFVHSAPWSVGDQGKRDV
MAV_4986|M.avium_104 MDSSTYGVPVNST--YGYKVTVEDAVRFDNVGDYVHSAPWSVDDQGKRDV
Mflv_0479|M.gilvum_PYR-GCK MDSSTYGVPVDDPSGEGYKLKVKDAVRIDNSGIFVHGAPWSVGSQGESNV
Mvan_0177|M.vanbaalenii_PYR-1 MDSSTYGVPVDDPSGEGYKLKVQDAVRIDNSGIFVHSAPWSVGAQGESNV
TH_3385|M.thermoresistible__bu MDSSTYGVPVDDP--EGYKIKVKDAVRIDNSGIFVHGAPWSTAAQGSRNV
MSMEG_0233|M.smegmatis_MC2_155 MDSSTYGVPVDSA--EGYKLKVKDAVRIDNSGIFVHSAPWSVGDQGKRNV
MAB_4537c|M.abscessus_ATCC_199 MDSSTYGVPVNSA--EGYKLDVYWATRLSNSGIFVHAAPWSVGAQGKSDT
MLBr_00426|M.leprae_Br4923 MDSSTYGVPVNSP--NGYRADVDWATQLSYSGVFLHSAPWSVGAQGHTNT
**********:.. **: * *.::. * ::*.****. ** :.
Mb0198|M.bovis_AF2122/97 THGCINLSPANAKWFYDNFGSGDPVVVKNSVG-TYNKNDGAQDWQI----
Rv0192|M.tuberculosis_H37Rv THGCINLSPANAKWFYDNFGSGDPVVVKNSVG-TYNKNDGAQDWQI----
MMAR_0435|M.marinum_M SHGCINLSPTNAKWFFDNFGSGDPIVVKNSVG-TYNKNDGAQDWQI----
MUL_1085|M.ulcerans_Agy99 SHGCINLSPTNAKWFFDNFGSGDPIVVKNSVG-TYNKNDGAQDWQI----
MAV_4986|M.avium_104 SHGCINISPANAKWFFDNFGPGDPIIVKNSSGGDYKKNDGSADWMN----
Mflv_0479|M.gilvum_PYR-GCK SHGCINLSAENAKWFRENFGSGDAVVIKNTDGGLYTQPDGASDWQMF---
Mvan_0177|M.vanbaalenii_PYR-1 SHGCINLSPANAQWFYDNFGSGDAVVIKNTDGGMYTQPDGASDWQMF---
TH_3385|M.thermoresistible__bu SHGCINLSPEDAEWFFDTFGSGDPVVIKNTEGGWYTQQDGAHDWQLF---
MSMEG_0233|M.smegmatis_MC2_155 SHGCINLSPENARWFYEQFGSGDPVVIKNSKGGWYDQPDGASDWQMF---
MAB_4537c|M.abscessus_ATCC_199 SHGCINVNTDNATWFFNQSHPGDPVIVKNSPGGPYKDYDGYDDWQRF---
MLBr_00426|M.leprae_Br4923 SHGCLNVSPSNAQWFYDHVKRGDIVEVVNTVGDTLPGAEGLGDWNIPWEQ
:***:*:.. :* ** : ** : : *: * :* **
Mb0198|M.bovis_AF2122/97 --------
Rv0192|M.tuberculosis_H37Rv --------
MMAR_0435|M.marinum_M --------
MUL_1085|M.ulcerans_Agy99 --------
MAV_4986|M.avium_104 --------
Mflv_0479|M.gilvum_PYR-GCK --------
Mvan_0177|M.vanbaalenii_PYR-1 --------
TH_3385|M.thermoresistible__bu --------
MSMEG_0233|M.smegmatis_MC2_155 --------
MAB_4537c|M.abscessus_ATCC_199 --------
MLBr_00426|M.leprae_Br4923 WKAGNANI