For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGWTRGTLFAALNAAVVSVVGLALVLSAGPALADPDPAPADPGAVAAPPGPPAPPDPLAPPPPPDPLAPP PPAAPPAPWLPPAAQPAAAPAAGQDPTPFTGTPPFGPPTFVPKTGSTVGVAQPIIINFPGRVDDAGAAIS AVHVSSVPPVPGKFYWMTPTQLRWRPLSFWPAHTAVTVDAGGTVTNFQTGDTLVATADDATHQLTVTRNG TVEKTFPMSMGMTAGNHQTPNGTYYVQDKKASVVMDSSTYGVPVNSTYGYKVTVEDAVRFDNVGDYVHSA PWSVDDQGKRDVSHGCINISPANAKWFFDNFGPGDPIIVKNSSGGDYKKNDGSADWMN*
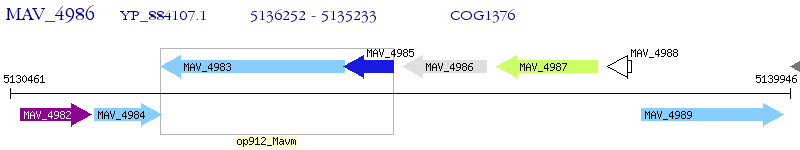
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4986 | - | - | 100% (339) | ErfK/YbiS/YcfS/YnhG family protein |
| M. avium 104 | MAV_1661 | - | 3e-50 | 40.23% (261) | ErfK/YbiS/YcfS/YnhG family protein |
| M. avium 104 | MAV_5194 | - | 5e-49 | 45.87% (218) | ErfK/YbiS/YcfS/YnhG family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0198 | - | 1e-136 | 67.34% (349) | hypothetical protein Mb0198 |
| M. gilvum PYR-GCK | Mflv_2824 | - | 2e-97 | 58.45% (296) | ErfK/YbiS/YcfS/YnhG family protein |
| M. tuberculosis H37Rv | Rv0192 | - | 1e-124 | 68.65% (303) | hypothetical protein Rv0192 |
| M. leprae Br4923 | MLBr_00426 | lppS | 1e-50 | 42.39% (243) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_4537c | - | 9e-92 | 53.68% (326) | hypothetical protein MAB_4537c |
| M. marinum M | MMAR_0435 | - | 1e-137 | 69.57% (345) | secretory protein |
| M. smegmatis MC2 155 | MSMEG_0233 | - | 1e-104 | 56.29% (318) | lipoprotein Lpps |
| M. thermoresistible (build 8) | TH_3385 | - | 8e-87 | 51.40% (286) | PUTATIVE ErfK/YbiS/YcfS/YnhG family protein |
| M. ulcerans Agy99 | MUL_1085 | - | 1e-134 | 70.03% (337) | secretory protein |
| M. vanbaalenii PYR-1 | Mvan_3694 | - | 2e-99 | 56.01% (316) | ErfK/YbiS/YcfS/YnhG family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0198|M.bovis_AF2122/97 ---MSRWKQGWTRG-SLFAALNIAAVVAV---LMLGAGVAVADPDAAPGD
Rv0192|M.tuberculosis_H37Rv ---MPHWAEERHRRESNYVALEAGLDEGE---SIRRSEHSRSGCGADAGC
MMAR_0435|M.marinum_M ---MSRWTRGRIRG-SLFAAVNAAGVVGV---LLLGAGPAVADPDDAPGD
MUL_1085|M.ulcerans_Agy99 ---MSRWMRGRIRG-SLFAAVNAAGVVGV---LLLGAGPAVADPDDAPGD
MAV_4986|M.avium_104 ---MSGWTRG-----TLFAALNAAVVSVVGLALVLSAGPALADPDPAPAD
Mflv_2824|M.gilvum_PYR-GCK ---MRCSPAR-----QILAMMTAALAAVT----VLTAPSASAEP------
Mvan_3694|M.vanbaalenii_PYR-1 ---MRCPPPR-----PFMAMMTAALAAVT----VLTAPSASAEP------
MSMEG_0233|M.smegmatis_MC2_155 ---MPKSAKR-----RLITAFMAAGLVGG----LVMSPSASADP------
TH_3385|M.thermoresistible__bu --------------------------------------------------
MAB_4537c|M.abscessus_ATCC_199 MLVLTRWFRR-----ALVSGVCVAGLLGA------GVVPALADP------
MLBr_00426|M.leprae_Br4923 MGTATRRVQPKAWRALLTLLVISVVMPGVACNRGGGNVPVNVIGDKGTPF
Mb0198|M.bovis_AF2122/97 P---GGPG----------GPGGTAGPVDPPAVDLLAPPPDPLALPPALDP
Rv0192|M.tuberculosis_H37Rv WRCRGGPGRGSRRSRRSRGPGGTAGPVDPPAVDLLAPPPDPLALPPALDP
MMAR_0435|M.marinum_M P--------------------PVIAPAEP-APDPLAPPPGPLALPPMPDP
MUL_1085|M.ulcerans_Agy99 P--------------------PVIAPAEP-APDPLAPPPGPLALPPMPDP
MAV_4986|M.avium_104 P-------------------GAVAAPPGP------PAPPDPLAPPPPPDP
Mflv_2824|M.gilvum_PYR-GCK ---------------------QIPVP-APGDPLTVPDSAVPNPFPPPVSG
Mvan_3694|M.vanbaalenii_PYR-1 ---------------------QIPVP-TPGNPLTVPDSTAPNPFLPPLSG
MSMEG_0233|M.smegmatis_MC2_155 ---------------------EVPAPPVPVEPAAPPAPPNPFAFPPPAAP
TH_3385|M.thermoresistible__bu ------------------------LPPDHPAAVSAPAPFNPFAPQPTIPP
MAB_4537c|M.abscessus_ATCC_199 ---------------------GDPPDPGIEEPGPPPPPPLPWELPPPPPP
MLBr_00426|M.leprae_Br4923 ADLLVPKLTASVTDGAVGVNVDMPVTVTVADGVLAAVTMINDNGRMINGQ
. .
Mb0198|M.bovis_AF2122/97 LAPPP-PDPLAPPPPDP----------LAVPVAAGPVAGQDPTPFVGPPP
Rv0192|M.tuberculosis_H37Rv LAPPP-PDPLAPPPPDP----------LAVPVAAGPVAGQDPTSFVGPPP
MMAR_0435|M.marinum_M LAPPPFPDPLAPP--------------PLVPVAAGPVAGQDPTPFFGPPP
MUL_1085|M.ulcerans_Agy99 LAPP-----------------------PLVPVAAGPVAGQDPTPFFGPPP
MAV_4986|M.avium_104 LAPPPPAAPPAPWLP------------PAAQPAAAPAAGQDPTPFTGTPP
Mflv_2824|M.gilvum_PYR-GCK TFPGPSPTAT-----------------------EDTPAGQNAAPYLGPPV
Mvan_3694|M.vanbaalenii_PYR-1 PFPGPSSTAA-----------------------EQTPAGQNPAPYLGPPV
MSMEG_0233|M.smegmatis_MC2_155 AAVGPADPATPGATATDPDAQVVTNYGAPAVPIEGVPEGQNPEPFTGQPP
TH_3385|M.thermoresistible__bu ----------------------------------DTPAGQNPLPYTGEPV
MAB_4537c|M.abscessus_ATCC_199 PAPDAPPPAEVPP--------------VPAPVPTTPPPGAPPTGDVPPLP
MLBr_00426|M.leprae_Br4923 FSPDGLRWSTTEPLGFN----RCYTLSAKALGLGGVVNRQMTFQTSSPAH
.
Mb0198|M.bovis_AF2122/97 FRPPTFNPVDGAMVGVAKPIVINFAVPIADRAMAESAIHISSIPPVPGKF
Rv0192|M.tuberculosis_H37Rv FRPPTFNPVDGAMVGVAKPIVINFAVPIADRAMAESAIHISSIPPVPGKF
MMAR_0435|M.marinum_M FRPPSFNPVDGAMVGVAKPIIINFQVPIADQAMAESAIHISSIPPVPGKF
MUL_1085|M.ulcerans_Agy99 FRPPSFNPVDGAMVGVAKPIIINFQVPIADQAMAESAIHISSIPPVPGKF
MAV_4986|M.avium_104 FGPPTFVPKTGSTVGVAQPIIINFPGRVDDAGAAISAVHVSSVPPVPGKF
Mflv_2824|M.gilvum_PYR-GCK FAPPTFNPTNGSTVGVAKPIVINFQRPIADRTLAEQAVHISSTPAVPGKF
Mvan_3694|M.vanbaalenii_PYR-1 FAPPTFNPTNGSTVGVAKPIIINFQRPIADRALAEQAVHISSTPAVPGKF
MSMEG_0233|M.smegmatis_MC2_155 FLPPSFNPVNGSMVGVAKPIYINFQRPIANKQMAQDAIRITSNPPVPGRF
TH_3385|M.thermoresistible__bu FLPPSFNPVNGALVGVAKPISIKFQRPIANRKMAEDAIHISSEPPVPGRF
MAB_4537c|M.abscessus_ATCC_199 VGLLKNSPNNGDVVGVAQPITIVFAAPVTDKKAAEAAVKITTSKPAPGYF
MLBr_00426|M.leprae_Br4923 LTMPYVNPGNGEIVGIGEPVAIRFDENIANRLAAQKAITITANPPVEGAF
. * * **:.:*: * * : : * *: ::: .. * *
Mb0198|M.bovis_AF2122/97 YWMSPTQVRWRPFEFWPANTAVNIDAAG--------------TKSSFRTG
Rv0192|M.tuberculosis_H37Rv YWMSPTQVRWRPFEFWPANTAVNIDAAG--------------TKSSFRTG
MMAR_0435|M.marinum_M YWMTPTQVRWRPFQFWPANTAVNIDAAG--------------TKSSFRTG
MUL_1085|M.ulcerans_Agy99 YWMTPTQVRWRPFQFWPANTAVNIDAAG--------------TKSSFRTG
MAV_4986|M.avium_104 YWMTPTQLRWRPLSFWPAHTAVTVDAGG--------------TVTNFQTG
Mflv_2824|M.gilvum_PYR-GCK YWMSATQLRWRPLDFWPANTTVSIDASG--------------TKSSFRTG
Mvan_3694|M.vanbaalenii_PYR-1 YWMSATQLRWRPIDFWPANTTVTIDASG--------------TKSSFRTG
MSMEG_0233|M.smegmatis_MC2_155 YWVTDTQLRWRPQDFWPSNTVVNIDAAG--------------TKSSFRVG
TH_3385|M.thermoresistible__bu YWISDTEVRWRPQDFWPAHTVVNIDAGG--------------TKSSFRVG
MAB_4537c|M.abscessus_ATCC_199 YWYTDQQLRWKPTQFWPANTDVNVNAGG--------------TKWSFKVG
MLBr_00426|M.leprae_Br4923 YWLNNREVRWRPEHFWKSGTAVDVAVNTYGVDLGEGMFGEDNVKTHFTIG
** . ::**:* ** : * * : . . * *
Mb0198|M.bovis_AF2122/97 DSLVATADDATHQMTITRNGVVQKTFPMSMGMVSGGHQTPNGTYYVLEKF
Rv0192|M.tuberculosis_H37Rv DSLVATADDATHQMTITRNGVVQKTFPMSMGMVSGGHQTPNGTYYVLEKF
MMAR_0435|M.marinum_M DSLVATADDATHQMTITRNGVVEQTFPMSMGMAAGNHQTPNGTYYVLEKM
MUL_1085|M.ulcerans_Agy99 DSLVATADDATHQMTITRNGVVEQTFPMSMGMAAGNHQTPNGTYYVLEKM
MAV_4986|M.avium_104 DTLVATADDATHQLTVTRNGTVEKTFPMSMGMTAGNHQTPNGTYYVQDKK
Mflv_2824|M.gilvum_PYR-GCK DSLVATIDNATLQMEVMRNGTLEKTMPVSLGKPG--YDTPNGTYYVLEKF
Mvan_3694|M.vanbaalenii_PYR-1 DSLVATIDNATLQMEVMRNGTLEKTMPVSLGKPG--YDTPNGTYYVLEKF
MSMEG_0233|M.smegmatis_MC2_155 DYLVATVDDSTHQMEIRRNGKLEKTFPVSMGKNDGKHETKNGTYYVLEKF
TH_3385|M.thermoresistible__bu ESQVATIDDKTKQMEVVINGELVKTMPVSLGKKG--YETKNGTYYVLEKF
MAB_4537c|M.abscessus_ATCC_199 DAFVSTVDDATYTMTVTRNGVVERTMPISMGKHDKKHETKNGTYYVSEKF
MLBr_00426|M.leprae_Br4923 DEVFATADDATKMLTVRVNGEVVKIMPTSMGKDS--TPTANGIYIVGARF
: .:* *: * : : ** : : :* *:* * ** * * :
Mb0198|M.bovis_AF2122/97 ATVVMDSSTYGVPVNSAQGYKLTVSDAVRIDNSGNFVHSAPWSVADQGKR
Rv0192|M.tuberculosis_H37Rv ATVVMDSSTYGVPVNSAQGYKLTVSDAVRIDNSGNFVHSAPWSVADQGKR
MMAR_0435|M.marinum_M PTVVMDSSTYGVPVNSPQGYKVTVADAVRIDNSGNFVHSAPWSVGDQGKR
MUL_1085|M.ulcerans_Agy99 PTVVMDSSTYGVPVNSPQGYKVTVADAVRIDNSGNFVHSAPWSVGDQGKR
MAV_4986|M.avium_104 ASVVMDSSTYGVPVNSTYGYKVTVEDAVRFDNVGDYVHSAPWSVDDQGKR
Mflv_2824|M.gilvum_PYR-GCK ADMVMDSSTYGVPIDAAEGYKLKVKDAVRINNAGIFVHGAPWSVADQGKR
Mvan_3694|M.vanbaalenii_PYR-1 ADMVMDSSTYGVPVDAAEGYKLKVQDAVRINNAGIFVHGAPWSVADQGKR
MSMEG_0233|M.smegmatis_MC2_155 EDIVMDSSTYGVPVDSAEGYKLKVKDAVRIDNSGIFVHSAPWSVGDQGKR
TH_3385|M.thermoresistible__bu KDIVMDSSTYGVPVDDPEGYKIKVKDAVRIDNSGIFVHGAPWSTAAQGSR
MAB_4537c|M.abscessus_ATCC_199 QKMVMDSSTYGVPVNSAEGYKLDVYWATRLSNSGIFVHAAPWSVGAQGKS
MLBr_00426|M.leprae_Br4923 KHIIMDSSTYGVPVNSPNGYRADVDWATQLSYSGVFLHSAPWSVGAQGHT
::*********:: . **: * *.::. * ::*.****. **
Mb0198|M.bovis_AF2122/97 NVTHGCINLSPANAKWFYDNFGSGDPVVVKNSVG-TYNKNDGAQDWQI--
Rv0192|M.tuberculosis_H37Rv NVTHGCINLSPANAKWFYDNFGSGDPVVVKNSVG-TYNKNDGAQDWQI--
MMAR_0435|M.marinum_M DVSHGCINLSPTNAKWFFDNFGSGDPIVVKNSVG-TYNKNDGAQDWQI--
MUL_1085|M.ulcerans_Agy99 DVSHGCINLSPTNAKWFFDNFGSGDPIVVKNSVG-TYNKNDGAQDWQI--
MAV_4986|M.avium_104 DVSHGCINISPANAKWFFDNFGPGDPIIVKNSSGGDYKKNDGSADWMN--
Mflv_2824|M.gilvum_PYR-GCK NVSHGCPNLSPANAQWFYDNFGSGDPVVVKNSVG-IYDENDGAQDWQI--
Mvan_3694|M.vanbaalenii_PYR-1 NVSHGCPNLSPANAQWFYDNFGSGDPVVVKNSIG-IYDENDGAQDWQI--
MSMEG_0233|M.smegmatis_MC2_155 NVSHGCINLSPENARWFYEQFGSGDPVVIKNSKGGWYDQPDGASDWQMF-
TH_3385|M.thermoresistible__bu NVSHGCINLSPEDAEWFFDTFGSGDPVVIKNTEGGWYTQQDGAHDWQLF-
MAB_4537c|M.abscessus_ATCC_199 DTSHGCINVNTDNATWFFNQSHPGDPVIVKNSPGGPYKDYDGYDDWQRF-
MLBr_00426|M.leprae_Br4923 NTSHGCLNVSPSNAQWFYDHVKRGDIVEVVNTVGDTLPGAEGLGDWNIPW
:.:*** *:.. :* **:: ** : : *: * :* **
Mb0198|M.bovis_AF2122/97 ----------
Rv0192|M.tuberculosis_H37Rv ----------
MMAR_0435|M.marinum_M ----------
MUL_1085|M.ulcerans_Agy99 ----------
MAV_4986|M.avium_104 ----------
Mflv_2824|M.gilvum_PYR-GCK ----------
Mvan_3694|M.vanbaalenii_PYR-1 ----------
MSMEG_0233|M.smegmatis_MC2_155 ----------
TH_3385|M.thermoresistible__bu ----------
MAB_4537c|M.abscessus_ATCC_199 ----------
MLBr_00426|M.leprae_Br4923 EQWKAGNANI