For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAEAHCPGPAPQGPDRDTGVPTGPIPGSSKCYRDPVGAPGGRVPFRRVHLSGGRHLDLYDTSGPYTETDT PARLATGLPSRPGVVTDRGTQLQRAREGVVTSEMAFVAARESLRPKVVRDAVAAGHAVIPANHRHPESEP MIIGRAFRVKVNAGIGSPPDAVIGDMVRAIRWGADIVTDLSGSDCDRMVRNMPVPVGTVPLYQALGRVHG DIAALTWNTFCDTVIEQAESGVDCMTVHAGLRREHLSLAAGRITGIVSRGGAVIADWCLLHNAESFLYAH FDQLCETLARYDVTLALAAGLQPGSVADANDAAQIAELRTLGQLARVAHRHGVQVVVEGPGRVPMHKIAE SVRIHQQLCRNAPSYTIGPLTTDVAPEHDHATAVIGAAVAAQAGTSMLCCLPRPARLRESVIAHKIAAHS ADLAKGHPQAHEHDHAISRARCETHCRDRLALSPDPDAAFTGHDHSKGSYLPFP*
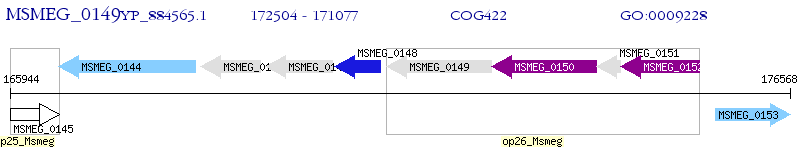
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0149 | - | - | 100% (475) | thiamine biosynthesis protein ThiC |
| M. smegmatis MC2 155 | MSMEG_0826 | thiC | e-154 | 59.91% (469) | thiamine biosynthesis protein ThiC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0431c | thiC | 1e-155 | 59.32% (472) | thiamine biosynthesis protein ThiC |
| M. gilvum PYR-GCK | Mflv_0754 | - | 1e-153 | 59.78% (465) | thiamine biosynthesis protein ThiC |
| M. tuberculosis H37Rv | Rv0423c | thiC | 1e-155 | 59.32% (472) | thiamine biosynthesis protein ThiC |
| M. leprae Br4923 | MLBr_00294 | thiC | 1e-152 | 58.39% (471) | thiamine biosynthesis protein ThiC |
| M. abscessus ATCC 19977 | MAB_4196 | - | 1e-152 | 58.25% (479) | thiamine biosynthesis protein ThiC |
| M. marinum M | MMAR_0735 | thiC | 1e-154 | 58.72% (470) | thiamine biosynthesis protein ThiC |
| M. avium 104 | MAV_4731 | thiC | 1e-156 | 59.62% (468) | thiamine biosynthesis protein ThiC |
| M. thermoresistible (build 8) | TH_1215 | thiC | 1e-157 | 59.57% (465) | PROBABLE THIAMINE BIOSYNTHESIS PROTEIN THIC |
| M. ulcerans Agy99 | MUL_3600 | thiC | 1e-154 | 58.72% (470) | thiamine biosynthesis protein ThiC |
| M. vanbaalenii PYR-1 | Mvan_0725 | - | 1e-146 | 57.85% (465) | thiamine biosynthesis protein ThiC |
CLUSTAL 2.0.9 multiple sequence alignment
TH_1215|M.thermoresistible__bu ---------VSDQSVAINPTVTTGPIAGSTKVYRELEGV---PGARVPYR
Mvan_0725|M.vanbaalenii_PYR-1 ------------MSDVVN--VTTGPIAGSTKVYRN----------GVPFR
Mflv_0754|M.gilvum_PYR-GCK ---------MTAVSDTVS--VTTGPITGSSKVYRELSD-----GGRVPFR
Mb0431c|M.bovis_AF2122/97 ------------MTITVEPSVTTGPIAGSAKAYREIEAPGSGATLQVPFR
Rv0423c|M.tuberculosis_H37Rv ------------MTITVEPSVTTGPIAGSAKAYREIEAPGSGATLQVPFR
MMAR_0735|M.marinum_M ------------MTVTVEPSITTGPIAGSSKAYREVAGP-DGVTLRVPLR
MUL_3600|M.ulcerans_Agy99 ------------MTVTVEPSITTGPIAGSSKAYREVAGP-DGVTLRVPLR
MAV_4731|M.avium_104 ------------MTAIVEPSVTTGPIAGSSKVYRELDGV-PGA--RVPFR
MLBr_00294|M.leprae_Br4923 --------MTETLSKTTEPSVTTGPIPGSSKAYREVANPDGGPSLRVPFR
MAB_4196|M.abscessus_ATCC_1997 -------MSTPSSRSQAPETVTTGPIQGSEKIYQELPNG-----LRVPQR
MSMEG_0149|M.smegmatis_MC2_155 MSAEAHCPGPAPQGPDRDTGVPTGPIPGSSKCYRDPVGA---PGGRVPFR
:.**** ** * *:: ** *
TH_1215|M.thermoresistible__bu RVHLSNGEHLDLYDTSGPYTDPDAVIDLSAGLPPRPGVVRDRGTQLQRAR
Mvan_0725|M.vanbaalenii_PYR-1 RVNLTTGEHLDLYDTSGPYTDAGAVIDLEAGLPHR-RVTRNRGTQLQRAR
Mflv_0754|M.gilvum_PYR-GCK RVHLTTGDHLDLYDTSGPYTDDTAVIDLAAGLPERSGVVRDSGTQLMRAR
Mb0431c|M.bovis_AF2122/97 RVHLSTGDHFDLYDTSGPYTDTDTVIDLTAGLPHRPGVVRDRGTQLQRAR
Rv0423c|M.tuberculosis_H37Rv RVHLSTGDHFDLYDTSGPYTDTDTVIDLTAGLPHRPGVVRDRGTQLQRAR
MMAR_0735|M.marinum_M RVHLSTGADFDLYDTSGPYTDPNAVIDLAVGLPARPGLVRDRGTQLQRAR
MUL_3600|M.ulcerans_Agy99 RVHLSTGADFDLYDTSGPYTDPNAVIDLAVGLPARPGLVRDRGTQLQRAR
MAV_4731|M.avium_104 RVHLSTGDHFDLYDTSGPYTDPDATIDLTAGLPARPGRVRDRGTQLQRAR
MLBr_00294|M.leprae_Br4923 RVHLSTGAHFDLYDTSGPYTDPDAVINLTAGLPPRPGVIRDRGTQLQRAR
MAB_4196|M.abscessus_ATCC_1997 RVNLTNGEYLDLYDTSGPYTDTNAVIDLHKGLPPRAGIVTDRGTQLQRAR
MSMEG_0149|M.smegmatis_MC2_155 RVHLSGGRHLDLYDTSGPYTETDTPARLATGLPSRPGVVTDRGTQLQRAR
**:*: * :**********: : * *** * : **** ***
TH_1215|M.thermoresistible__bu AGEITAEMAFIAEREGVPAELVRREVAAGRAVIPANHNHPEAEPMIIGKA
Mvan_0725|M.vanbaalenii_PYR-1 AGEITAEMAFIAEREGVSPELVRDEVARGRAVIPANHNHPEAEPMIIGKA
Mflv_0754|M.gilvum_PYR-GCK KGEITAEMAFIAEREGLPVELVRDEVARGRAVIPANHNHPESEPMIIGKA
Mb0431c|M.bovis_AF2122/97 AGEITAEMAFIAAREDMSAELVRDEVARGRAVIPANHHHPESEPMIIGKA
Rv0423c|M.tuberculosis_H37Rv AGEITAEMAFIAAREDMSAELVRDEVARGRAVIPANHHHPESEPMIIGKA
MMAR_0735|M.marinum_M AGEITAEMAFIAAREGMSAELVRDEVALGRAVIPANHNHPESEPMVIGKA
MUL_3600|M.ulcerans_Agy99 AGEITAEMAFIAAREGMSAELVRDEVALGRAVIPANHNHPESEPMVIGKA
MAV_4731|M.avium_104 AGEITAEMAFIAAREGMPAELVRDEVARGRAVIPANHNHPEIEPMIIGKA
MLBr_00294|M.leprae_Br4923 AGEITAEMAFIADREGMPAELVRVEVALGRAVIPANHNHPEIEPMIIGKA
MAB_4196|M.abscessus_ATCC_1997 AGEITAEMEFIAVREGVPAELVRTEVAAGRAVIPANHKHPESEPMIIGKA
MSMEG_0149|M.smegmatis_MC2_155 EGVVTSEMAFVAARESLRPKVVRDAVAAGHAVIPANHRHPESEPMIIGRA
* :*:** *:* **.: ::** ** *:*******.*** ***:**:*
TH_1215|M.thermoresistible__bu FSVKVNANIGNSAVTSSIEEEVDKMVWATRWGADTVMDLSTGRNIHETRE
Mvan_0725|M.vanbaalenii_PYR-1 FAVKINANIGNSAVTSSIAEEVDKMVWATRWGADTIMDLSTGRDIHQTRE
Mflv_0754|M.gilvum_PYR-GCK FAVKVNANIGNSAVTSSVAEEVDKMVWATRWGADTIMDLSTGADIHLTRE
Mb0431c|M.bovis_AF2122/97 FAVKVNANIGNSAVTSSIAEEVDKMVWATRWGADTIMDLSTGKNIHETRE
Rv0423c|M.tuberculosis_H37Rv FAVKVNANIGNSAVTSSIAEEVDKMVWATRWGADTIMDLSTGKNIHETRE
MMAR_0735|M.marinum_M FAVKVNANIGNSAVTSSIAEEVDKMVWATRWGADTIMDLSTGKNIHETRE
MUL_3600|M.ulcerans_Agy99 FAVKVNANIGNSAVTSSIAEEVDKMVWATRWGADTIMDLSTGKNIHETRE
MAV_4731|M.avium_104 FATKVNANIGNSAVTSSIAEEVDKMVWATRWGADTIMDLSTGKNIHETRE
MLBr_00294|M.leprae_Br4923 FAVKVNANIGNSAVTSSIAEEIDKMVWATRWGADTIMDLSTGKNIHETRE
MAB_4196|M.abscessus_ATCC_1997 FGVKINANIGNSAVTSSIAEEVEKMVWAIRWGADNIMDLSTGKDIHQTRE
MSMEG_0149|M.smegmatis_MC2_155 FRVKVNAGIG-----SPPDAVIGDMVRAIRWGADIVTDLSG-----SDCD
* .*:**.** *. : .** * ***** : *** :
TH_1215|M.thermoresistible__bu WILRNSPVPVGTVPIYQALEKVDGDPVKLTWEVYRDTVIEQCEQGVDYMT
Mvan_0725|M.vanbaalenii_PYR-1 WILRNSPVPVGTVPMYQALEKVGGDPVKLTWEVYRDTVIEQCEQGVDYMT
Mflv_0754|M.gilvum_PYR-GCK WILRNSPVPVGTVPIYQALEKVDGDPTQLTWKVYRDTVIEQCEQGVDYMT
Mb0431c|M.bovis_AF2122/97 WILRNSPVPVGTVPIYQALEKVKGDPTELTWEIYRDTVIEQCEQGVDYMT
Rv0423c|M.tuberculosis_H37Rv WILRNSPVPVGTVPIYQALEKVKGDPTELTWEIYRDTVIEQCEQGVDYMT
MMAR_0735|M.marinum_M WILRNSPVPVGTVPIYQALEKVKGDPTELTWELYRDTVIEQCEQGVDYMT
MUL_3600|M.ulcerans_Agy99 WILRNSPVPVGTVPIYQALEKVKGDPTELTWELYRDTVIEQCEQGVDYMT
MAV_4731|M.avium_104 WILRNSPVPVGTVPIYQALEKVKGDPTLLTWEIYRDTVIEQCEQGVDYMT
MLBr_00294|M.leprae_Br4923 WILRNSPVPVGTVPIYQALEKVKGDPTKLTWEIYRDTVIEQCEQGVDYMT
MAB_4196|M.abscessus_ATCC_1997 WILRNSPVPVGTVPIYQALEKTNGDPAALTWELYRDTVIEQAEQGVDYMT
MSMEG_0149|M.smegmatis_MC2_155 RMVRNMPVPVGTVPLYQALGRVHGDIAALTWNTFCDTVIEQAESGVDCMT
::** ********:**** :. ** . ***: : ******.*.*** **
TH_1215|M.thermoresistible__bu VHAGVLLRYIPLTVDRVTGIVSRGGSIMAAWCLAHHQESFLYTHFEELCE
Mvan_0725|M.vanbaalenii_PYR-1 VHAGVLLRHIPLTVDRVTGIVSRGGSIMAAWCLAHHRESFLYTNFEELCG
Mflv_0754|M.gilvum_PYR-GCK VHAGVLLRYIPLTVNRVTGIVSRGGSIMAAWMLAHHTESFLYTHFAELCE
Mb0431c|M.bovis_AF2122/97 VHAGVLLRYVPLTAKRVTGIVSRGGSIMAAWCLAHHRESFLYTNFEELCD
Rv0423c|M.tuberculosis_H37Rv VHAGVLLRYVPLTAKRVTGIVSRGGSIMAAWCLAHHRESFLYTNFEELCD
MMAR_0735|M.marinum_M VHAGVLLRYVPLTAKRVTGIVSRGGSIMAAWCLAHHRESFLYTNFEELCD
MUL_3600|M.ulcerans_Agy99 VHAGVLLRYVPLTAKRVTGIVSRGGSIMAAWCLAHHRESFLYTNFEELCD
MAV_4731|M.avium_104 VHAGVLLRYVPLTAKRVTGIVSRGGSIMAAWCLAHHRESFLYTNFDELCE
MLBr_00294|M.leprae_Br4923 VHAGVLLRYVLLTAKRVTGIVSRGGSIMASWCLANHRESFLYTNFAELCD
MAB_4196|M.abscessus_ATCC_1997 VHAGVLLRYVPLTAKRVTGIVSRGGSIMAAWCLAHHRESFLYTHFEELCE
MSMEG_0149|M.smegmatis_MC2_155 VHAGLRREHLSLAAGRITGIVSRGGAVIADWCLLHNAESFLYAHFDQLCE
****: .:: *:. *:********:::* * * :: *****::* :**
TH_1215|M.thermoresistible__bu ILARYDVTFSLGDGLRPGSIADANDEAQFAELRTLGELTKIAKSHGAQVM
Mvan_0725|M.vanbaalenii_PYR-1 ILARYDVTFSLGDGLRPGSIADANDAAQFAELRTLGELTGMAKSHGVQVM
Mflv_0754|M.gilvum_PYR-GCK ILARYDVTFSLGDGLRPGSIADANDEAQFAELRTLGELTKIAKAHGVQVM
Mb0431c|M.bovis_AF2122/97 IFARYDVTFSLGDGLRPGSIADANDAAQFAELRTLGELTKIAKAHGAQVM
Rv0423c|M.tuberculosis_H37Rv IFARYDVTFSLGDGLRPGSIADANDAAQFAELRTLGELTKIAKAHGAQVM
MMAR_0735|M.marinum_M ILARYDVTFSLGDGLRPGSIADANDAAQFAELRTLGELTKIAKAHGVQVM
MUL_3600|M.ulcerans_Agy99 ILARYDVTFSLGDGLRPGSIADANDAAQFAELRTLGELTKIAKAHGVQVM
MAV_4731|M.avium_104 IFARYDVTFSLGDGLRPGSIADANDAAQFAELRTLGELTKIAKSHGVQVM
MLBr_00294|M.leprae_Br4923 IFARYDVTFSLGDGLRPGSIADANDTAQFAELRTLGELSKIAKVHGAQVM
MAB_4196|M.abscessus_ATCC_1997 ILARYDVTFSLGDGLRPGSIADANDEAQFAELRTLGELTKIAKSHGVQVM
MSMEG_0149|M.smegmatis_MC2_155 TLARYDVTLALAAGLQPGSVADANDAAQIAELRTLGQLARVAHRHGVQVV
:******::*. **:***:***** **:*******:*: :*: **.**:
TH_1215|M.thermoresistible__bu IEGPGHVPMHKIAENVRLEEEWCEEAPFYTLGPLATDIAPGYDHITSAIG
Mvan_0725|M.vanbaalenii_PYR-1 IEGPGHVPMHKIVENVRLEEELCEEAPFYTLGPLATDIAPAYDHITSAIG
Mflv_0754|M.gilvum_PYR-GCK IEGPGHVPMHKIVENVRLEEELCEEAPFYTLGPLTTDIAPAYDHITSAIG
Mb0431c|M.bovis_AF2122/97 IEGPGHIPMHKIVENVRLEEELCEEAPFYTLGPLATDIAPAYDHITSAIG
Rv0423c|M.tuberculosis_H37Rv IEGPGHIPMHKIVENVRLEEELCEEAPFYTLGPLATDIAPAYDHITSAIG
MMAR_0735|M.marinum_M IEGPGHVPMHKIVENVRLEEELCEEAPFYTLGPLATDIAPAYDHITSAIG
MUL_3600|M.ulcerans_Agy99 IEGPGHVPMHKIVENVRLEEELCEEAPFYTLGPLATDIAPAYDHITSAIG
MAV_4731|M.avium_104 IEGPGHVPMHKIVENVRLEEEWCEEAPFYTLGPLATDIAPAYDHITSAIG
MLBr_00294|M.leprae_Br4923 IEGPGHIPMHKIVENVRLEEELCEEAPFYTLGPLATDIAPAYDHITSAIG
MAB_4196|M.abscessus_ATCC_1997 IEGPGHVPMHKIVENVKLEEELCEEAPFYTLGPLATDIAPAYDHITSAIG
MSMEG_0149|M.smegmatis_MC2_155 VEGPGRVPMHKIAESVRIHQQLCRNAPSYTIGPLTTDVAPEHDHATAVIG
:****::*****.*.*::.:: *.:** **:***:**:** :** *:.**
TH_1215|M.thermoresistible__bu ASIIAQAGTAMLCYVTPKEHLGLPNRQDVKDGVIAYKIAAHAADLAKGHP
Mvan_0725|M.vanbaalenii_PYR-1 AAIIAQAGTAMLCYVTPKEHLGLPNRKDVKDGVIAYKIAAHAADLAKGHP
Mflv_0754|M.gilvum_PYR-GCK AAIIAQAGTAMLCYVTPKEHLGLPDRSDVKTGVITYKIAAHAADLAKAHP
Mb0431c|M.bovis_AF2122/97 AAIIAQAGTAMLCYVTPKEHLGLPDRKDVKDGVIAYKIAAHAADLAKGHP
Rv0423c|M.tuberculosis_H37Rv AAIIAQAGTAMLCYVTPKEHLGLPDRKDVKDGVIAYKIAAHAADLAKGHP
MMAR_0735|M.marinum_M AAIIAQAGTAMLCYVTPKEHLGLPDRKDVKDGVIAYKIAAHAGDLAKGHP
MUL_3600|M.ulcerans_Agy99 AAIIAQAGTAMLCYVTPKEHLGLPDRKDVKDGVIAYKIAAHAGDLAKGHP
MAV_4731|M.avium_104 AAIIAQAGTAMLCYVTPKEHLGLPDRKDVKDGVIAYKIAAHAGDLAKGHP
MLBr_00294|M.leprae_Br4923 AAVIAQAGTAMLCYVTPKEHLGLPDRKDVKDGVIAYKIAAHAADLAKGYP
MAB_4196|M.abscessus_ATCC_1997 AAIIAQAGTAMLCYVTPKEHLGLPDRKDVKDGVIAYKIAAHSADLAKGHP
MSMEG_0149|M.smegmatis_MC2_155 AAVAAQAGTSMLCCLP------RPAR--LRESVIAHKIAAHSADLAKGHP
*:: *****:*** :. * * :: .**::*****:.****.:*
TH_1215|M.thermoresistible__bu RAQQRDDALSKARFEFRWHDQFALSLDPDTAREYHDETLPAEPAKTAHFC
Mvan_0725|M.vanbaalenii_PYR-1 GAQARDDALSKARFEFRWEDQFNLSLDPDTAREFHDETLPATPAKTAHFC
Mflv_0754|M.gilvum_PYR-GCK RAQERDDALSTARFEFRWNDQFALSLDPDSACGFHDETLPAVPAKTAHFC
Mb0431c|M.bovis_AF2122/97 RAQERDDALSTARFEFRWNDQFALSLDPDTAREFHDETLPAEPAKTAHFC
Rv0423c|M.tuberculosis_H37Rv RAQERDDALSTARFEFRWNDQFALSLDPDTAREFHDETLPAEPAKTAHFC
MMAR_0735|M.marinum_M HAQERDNALSQARFEFRWNDQFALSLDPDTAREYHDETLPAEPAKTAHFC
MUL_3600|M.ulcerans_Agy99 HAQERDNALSQARFEFRWNDQFALSLDPDTAREYHDETLPAEPAKTAHFC
MAV_4731|M.avium_104 HAQERDNALSQARFEFRWNDQFALSLDPDTAREYHDETLPAEPAKTAHFC
MLBr_00294|M.leprae_Br4923 RAQERDDALSTARFEFRWNDQFALSLDPPTAREFHDETLPAEPAKTAHFC
MAB_4196|M.abscessus_ATCC_1997 RAQLRDNALSKARFEFRWEDQFNLSLDPDTAREFHDETLPAEPAKTAHFC
MSMEG_0149|M.smegmatis_MC2_155 QAHEHDHAISRARCETHCRDRLALSPDPDAAFTGHDHSK-----------
*: :*.*:* ** * : .*:: ** ** :* **.:
TH_1215|M.thermoresistible__bu SMCGPRFCSMRITADIRDA-------------------MAQKSREFAEHG
Mvan_0725|M.vanbaalenii_PYR-1 SMCGPKFCSMRISHEVQEA-------------------MKEKSREFADQG
Mflv_0754|M.gilvum_PYR-GCK SMCGPKFCSMRISRDVRDAYEKG---------------MTDKSQEFAKHG
Mb0431c|M.bovis_AF2122/97 SMCGPKFCSMRITQDVREYAAEHGLETEADIEAVLAAGMAEKSREFAEHG
Rv0423c|M.tuberculosis_H37Rv SMCGPKFCSMRITQDVREYAAEHGLETEADIEAVLAAGMAEKSREFAEHG
MMAR_0735|M.marinum_M SMCGPKFCSMRITQDVRDYAAKHGLDSEEAIEAALEAGMAEKSAEFADHG
MUL_3600|M.ulcerans_Agy99 SMCGPKFCSMRITRDVRDYAAKHGLDSEEAIEAALEAGMAEKSAEFADHG
MAV_4731|M.avium_104 SMCGPKFCSMRITQDVRDYAAKHGLDSEEAIEAA----MADKSREFAEHG
MLBr_00294|M.leprae_Br4923 SMCGPKFCSMRITADIRVYAAKHGLDTEEAIEMG----MTEKSAEFAEHG
MAB_4196|M.abscessus_ATCC_1997 SMCGPKFCSMRITADIREFAAENGLETQEDIDAMLARGMEEKSAEFAEHG
MSMEG_0149|M.smegmatis_MC2_155 -------------------------------------------------G
*
TH_1215|M.thermoresistible__bu NQVYLPLEEVRP
Mvan_0725|M.vanbaalenii_PYR-1 NKVYLPVVTS--
Mflv_0754|M.gilvum_PYR-GCK NRIYLPLHP---
Mb0431c|M.bovis_AF2122/97 NRVYLPITQ---
Rv0423c|M.tuberculosis_H37Rv NRVYLPITQ---
MMAR_0735|M.marinum_M NRVYLPITQ---
MUL_3600|M.ulcerans_Agy99 NRVYLPITQ---
MAV_4731|M.avium_104 NRVYLPLAQ---
MLBr_00294|M.leprae_Br4923 NRVYLPLTQ---
MAB_4196|M.abscessus_ATCC_1997 NRVYLPIA----
MSMEG_0149|M.smegmatis_MC2_155 S--YLPFP----
. ***.