For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FRLENVATAAYVVAALLFILALAGLSKHETSRAGNTFGIAGMAVALIATIVLAIHGQIEPLGLGLLVVAM IVGALIGLWRAKVVEMTGMPELIALLHSFVGLAAVLVGWNGYLHVEADPGSDEALKLAAEGMSGIHSAEV FIGVFIGAVTFTGSIVANLKLSARIKSAPLMLPGKNLLNIGALIVFAVLTVWFVIDPQLWLLVVVTVLAL LLGWHLVASIGGGDMPVVVSMLNSYSGWAAAASGFLLGNDLLIITGALVGSSGAYLSYIMCKAMNRSFIS VIAGGFGIENTGAGSDVDYGEHREINAEGVAELLAHANSVIITPGYGMAVAQAQYGVADLTRKLRERGVN VRFGIHPVAGRLPGHMNVLLAEAKVPYDIVLEMDEINDDFDGTDVVLVIGANDTVNPAAAEDPGSPIAGM PVLQVWNADNVIVFKRSMASGYAGVQNPLFFRDNTQMLFGDARDRVNDILAALPAPERV*
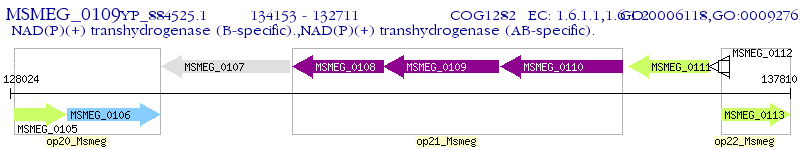
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0109 | pntB | - | 100% (480) | NAD(P) transhydrogenase, beta subunit |
| M. smegmatis MC2 155 | MSMEG_4108 | pntB | 0.0 | 95.00% (480) | NAD(P) transhydrogenase, beta subunit |
| M. smegmatis MC2 155 | MSMEG_0150 | - | e-103 | 42.80% (486) | NAD(P) transhydrogenase beta subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0162 | pntB | 3e-97 | 42.95% (475) | NAD(P) transhydrogenase subunit beta |
| M. gilvum PYR-GCK | Mflv_0727 | - | 0.0 | 87.08% (480) | NAD(P) transhydrogenase, beta subunit |
| M. tuberculosis H37Rv | Rv0157 | pntB | 3e-97 | 42.95% (475) | NAD(P) transhydrogenase subunit beta |
| M. leprae Br4923 | MLBr_02634 | pntB | 3e-94 | 41.83% (471) | pyridine transhydrogenase subunit beta |
| M. abscessus ATCC 19977 | MAB_4577c | - | 1e-95 | 41.03% (485) | NAD(P) transhydrogenase, beta subunit PntB |
| M. marinum M | MMAR_0379 | pntB | 7e-96 | 41.63% (478) | NAD(P) transhydrogenase (subunit beta) PntB |
| M. avium 104 | MAV_5140 | - | 1e-94 | 41.26% (475) | NAD(P) transhydrogenase beta subunit |
| M. thermoresistible (build 8) | TH_0162 | pntB | 1e-103 | 44.81% (482) | PROBABLE NAD(P) TRANSHYDROGENASE (SUBUNIT BETA) PNTB |
| M. ulcerans Agy99 | MUL_4733 | pntB | 9e-96 | 41.81% (476) | NAD(P) transhydrogenase (subunit beta) PntB |
| M. vanbaalenii PYR-1 | Mvan_0119 | - | 0.0 | 88.12% (480) | NAD(P) transhydrogenase, beta subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0379|M.marinum_M -----MNYLVIGLYILAFSLFIYGLMGLTGPKTAVRGNLIAAVGMAIAVA
MUL_4733|M.ulcerans_Agy99 -----MNYLVIGLYILAFSLFIYGLMGLTEPKTAVRGNLIAAVGMAIAVA
MAV_5140|M.avium_104 -----MNYLVIGLYIVSFALFIYGLMGLTGPKTAVRGNLIAAVGMAIAVA
MLBr_02634|M.leprae_Br4923 -----MNYFVIGLYILAFALFITGLMGLTGPRTAVSGNLIAAVGMTIAVA
Mb0162|M.bovis_AF2122/97 ---MNLHYLVEILYIISFSLFIYGLMGLTGPKTAVRGNLIAAAGMTIAVA
Rv0157|M.tuberculosis_H37Rv ---MNLHYLVEILYIISFSLFIYGLMGLTGPKTAVRGNLIAAAGMTIAVA
TH_0162|M.thermoresistible__bu -----MNYVVSILYIVSFAMFIYGLMGLTGPKTAVRGNWIAAAGMAVAII
MAB_4577c|M.abscessus_ATCC_199 MSSESLNYIVDVLYIAAFALFIYGLSGLTGPKTAVRGNWIAAVGMGIAVL
Mflv_0727|M.gilvum_PYR-GCK --MFTLETAATAAYVVAALLFILALAGLSKHETSRAGNTFGITGMAVALI
Mvan_0119|M.vanbaalenii_PYR-1 --MFTLETAATAAYVVAALLFILALAGLSRHETSRAGNTFGIVGMVVALV
MSMEG_0109|M.smegmatis_MC2_155 --MFRLENVATAAYVVAALLFILALAGLSKHETSRAGNTFGIAGMAVALI
:. . *: : :** .* **: .*: ** :. .** :*:
MMAR_0379|M.marinum_M ATLVKIRHTD----QWVLIIAGLVVGVALGVPPARLTKMTAMPQLVAFFN
MUL_4733|M.ulcerans_Agy99 ATLVKIRHTD----QWVLIIVGLVVGVALGVPPARLTKMTAMPQLVAFFN
MAV_5140|M.avium_104 ATLIKIRHTD----QWVLIIAGLVVGVVLGVPPARYTKMTAMPQLVAFFN
MLBr_02634|M.leprae_Br4923 ATLVKIRHTE----QWVLIIAALVVGIVLGFPPARLTKVTAMPQLVAFFN
Mb0162|M.bovis_AF2122/97 ATLVMIRHTS----QWPLIIAGLVVGVVLGVPPARLTKMTAMPQLVAFFN
Rv0157|M.tuberculosis_H37Rv ATLVMIRHTS----QWPLIIAGLVVGVVLGVPPARLTKMTAMPQLVAFFN
TH_0162|M.thermoresistible__bu ATLILIRDTD----NWPLIIAGLLIGVVLGVPPARMTKMTAMPQLVALFN
MAB_4577c|M.abscessus_ATCC_199 ATLIKVSESATT-LDWILIGAGLGIGVVLGIPPALKTKMTAMPQLVALFN
Mflv_0727|M.gilvum_PYR-GCK ATVALAIARDIEPLGLALLFAAMAIGAAIGLWRAKIVEMTGMPELIALLH
Mvan_0119|M.vanbaalenii_PYR-1 ATIALALARHIEPLGLALLVGAMTIGAAIGLWRAKIVEMTGMPELIALLH
MSMEG_0109|M.smegmatis_MC2_155 ATIVLAIHGQIEPLGLGLLVVAMIVGALIGLWRAKVVEMTGMPELIALLH
**: *: .: :* :*. * .::*.**:*:*:::
MMAR_0379|M.marinum_M GVGGGTVALIALSEFIETKG----FSAFQHGESPTVHIVVASLFA-AIIG
MUL_4733|M.ulcerans_Agy99 GVGGGTVALIALSEFIETKG----FSAFQHGESPTVHIVVASLFA-AIIG
MAV_5140|M.avium_104 GVGGGTVALIALSEFIETSG----FSAFQHGESPTVHIVVASLFA-AIIG
MLBr_02634|M.leprae_Br4923 GVGGGTVALIALSEFIETSG----FSEFQHHESPTVHIVVASLFA-AIIG
Mb0162|M.bovis_AF2122/97 GVGGGTVALIALSEFIDTTG----FSAFQHGESPTVHIVVASLFA-AIIG
Rv0157|M.tuberculosis_H37Rv GVGGGTVALIALSEFIDTTG----FSAFQHGESPTVHIVVASLFA-AIIG
TH_0162|M.thermoresistible__bu GVGGGTVALIALAEFMDTEG----FSAFKHGEEPTVHIVVGSLFA-AIIG
MAB_4577c|M.abscessus_ATCC_199 GVGGGTVALIAWSEFIETKG----FSEFKPDQSPTVALVVGSLFA-AIIG
Mflv_0727|M.gilvum_PYR-GCK SFVGLAAVLVGWNGYLHVEGDPGGAEAAHLASEGMLGIHSGEVVIGVFIG
Mvan_0119|M.vanbaalenii_PYR-1 SFVGLAAVLVGWNGYLHVEGDPGSAEALQLGSEGMLGIHSAEVVIGVFIG
MSMEG_0109|M.smegmatis_MC2_155 SFVGLAAVLVGWNGYLHVEADPGSDEALKLAAEGMSGIHSAEVFIGVFIG
.. * :..*:. ::.. . . : . : ..:. .:**
MMAR_0379|M.marinum_M SISFWGSIIAFGKLQELISG----APIGFGKAQQPVNLLLLVGAVVAAVV
MUL_4733|M.ulcerans_Agy99 SISFWGSIIAFGKLQELISG----APIGFGKAQQPVNLLLLVGAVVAAVV
MAV_5140|M.avium_104 SISFWGSIIAFGKLQEVISG----APIGFGKAQQPINLLLLAGAVAAAVV
MLBr_02634|M.leprae_Br4923 SISFWGSIIAFGKLQETISG----FPIGFGKVQQPINLLLLVGAIVAAVV
Mb0162|M.bovis_AF2122/97 SISFWGSIVAFGKLQEIISG----RPIGLGKAQQPINLLLLAVAVAAAVV
Rv0157|M.tuberculosis_H37Rv SISFWGSIVAFGKLQEIISG----RPIGLGKAQQPINLLLLAVAVAAAVV
TH_0162|M.thermoresistible__bu SISFWGSLIAFGKLQEILPG----RPIGFGKVQQPFNILLLIGSVAACVV
MAB_4577c|M.abscessus_ATCC_199 SVSFWGSLVAFAKLQESIPKNVEKRLVASAKLFQASNIVLLLAAVGTAVY
Mflv_0727|M.gilvum_PYR-GCK AVTFTGSIVANLKLSAKMKS---APMMLPGKNFLNVGALVVFAALTVWFV
Mvan_0119|M.vanbaalenii_PYR-1 AVTFTGSIVANLKLSARIKS---APLMLPGKNILNVGALVVFIALTVWFV
MSMEG_0109|M.smegmatis_MC2_155 AVTFTGSIVANLKLSARIKS---APLMLPGKNLLNIGALIVFAVLTVWFV
:::* **::* **. : : .* . ::: : . .
MMAR_0379|M.marinum_M IGLDAHPGSGGVSLWWMIGLLAAAGVLGLMVVLPIGGADMPVVISLLNAM
MUL_4733|M.ulcerans_Agy99 IGLDAYPGSGGVSLWWMIGLLAAAGVLGLMVVLPIGGADMPVVISLLNAM
MAV_5140|M.avium_104 IGLHAHPGSGGVSLWWMIGLLAAAGVLGLMVVLPIGGADMPVVISLLNAM
MLBr_02634|M.leprae_Br4923 IGLHAQPGSAGASLWWMIGLLVAAGVLGLMVVLSIGGADMPVVISLLNAM
Mb0162|M.bovis_AF2122/97 IGLHAHPGSGGVALWWMIGLLVAAGVLGLMVVLPIGGADMPVVISMLNAM
Rv0157|M.tuberculosis_H37Rv IGLHAHPGSGGVALWWMIGLLVAAGVLGLMVVLPIGGADMPVVISMLNAM
TH_0162|M.thermoresistible__bu IGLDAHPGTGGASLWWMIAVLVLAGVLGLMVVLPIGGADMPVVISMLNAL
MAB_4577c|M.abscessus_ATCC_199 IGLHP-----GLPVWWIVAVLVFAGVMGLFVVFPIGGADMPVVISLLNAM
Mflv_0727|M.gilvum_PYR-GCK I---------DPALWLLIVVTVLALLLGWHLVASIGGGDMPVVVSMLNSY
Mvan_0119|M.vanbaalenii_PYR-1 I---------DPQLWLLVVVTVLALALGWHLVASIGGGDMPVVVSMLNSY
MSMEG_0109|M.smegmatis_MC2_155 I---------DPQLWLLVVVTVLALLLGWHLVASIGGGDMPVVVSMLNSY
* . :* :: : . * :* :* .***.*****:*:**:
MMAR_0379|M.marinum_M TGLSAAAAGLALNNTAMIVAGMIVGASGSILTNLMAKAMNRSIPAIVAGG
MUL_4733|M.ulcerans_Agy99 TGLSAAAAGLALNNTAMIGAGMIVGASGSILTNLMAKAMNRSIPAIVAGG
MAV_5140|M.avium_104 TGLSAAAAGLALNNTAMIVAGMIVGASGSILTNLMAKAMNRSIPAIVAGG
MLBr_02634|M.leprae_Br4923 TGLSAAATGLALNNTAIIVAGMIVGASGSILTNLMAKAMNRSIPAIVAGG
Mb0162|M.bovis_AF2122/97 TGLSAAAAGLALNNTAMIVAGMIVGASGSILTNLMAKAMNRSIPAIVAGG
Rv0157|M.tuberculosis_H37Rv TGLSAAAAGLALNNTAMIVAGMIVGASGSILTNLMAKAMNRSIPAIVAGG
TH_0162|M.thermoresistible__bu TGLSAAAAGMALNNQAMIVAGMLVGASGSILTNLMAKAMNRSIPAIVAGG
MAB_4577c|M.abscessus_ATCC_199 TGLSAAAAGLALNNTAMIVAGMIVGASGSILTNLMAVAMNRSIPAIVFGS
Mflv_0727|M.gilvum_PYR-GCK SGWAAAAAGFLLGNDLLIVTGALVGSSGAYLSYIMCKAMNRSFISVIAGG
Mvan_0119|M.vanbaalenii_PYR-1 SGWAAAAAGFLLGNDLLIVTGALVGSSGAYLSYIMCKAMNRSFISVIAGG
MSMEG_0109|M.smegmatis_MC2_155 SGWAAAASGFLLGNDLLIITGALVGSSGAYLSYIMCKAMNRSFISVIAGG
:* :***:*: *.* :* :* :**:**: *: :*. *****: ::: *.
MMAR_0379|M.marinum_M FGGGGVSPSADGGGDKHVKATSAADAAIQMAYANQVIVVPGYGLAVAQAQ
MUL_4733|M.ulcerans_Agy99 FGGGGVSPSADGGGDKHVKATSAADAAIQMAYANQVIVVPGYGLAVAQAQ
MAV_5140|M.avium_104 FGGGGVAPGGGDGGDKHVKSTSAADAAIQMAYANQVIVVPGYGLAVAQAQ
MLBr_02634|M.leprae_Br4923 FGSGGVAPSRD--GDKQVKSTSAADAAIQMAYANQVIVVPGYGLAVAQAQ
Mb0162|M.bovis_AF2122/97 FGGGGVAPSGG-GDDKHVKATSAADAAIQMAYANQVIVVPGYGLAVAQAQ
Rv0157|M.tuberculosis_H37Rv FGGGGVAPSGG-GDDKHVKATSAADAAIQMAYANQVIVVPGYGLAVAQAQ
TH_0162|M.thermoresistible__bu FGGGGGVGGDGDSGEKVVKSTSAADAAIQMAYANQVIIVPGYGLAVAQAQ
MAB_4577c|M.abscessus_ATCC_199 FGGSDAGAAGGAAEQGTVKATSASDAAIQMAYANQVIVVPGYGLAVAQAQ
Mflv_0727|M.gilvum_PYR-GCK FGIE-AGPAED-KDYGEHREITADGAAELLASASSVIITPGYGMAVAQAQ
Mvan_0119|M.vanbaalenii_PYR-1 FGIE-AGPAED-KDYGEHREITAEGAAELLASASSVIITPGYGMAVAQAQ
MSMEG_0109|M.smegmatis_MC2_155 FGIENTGAGSD-VDYGEHREINAEGVAELLAHANSVIITPGYGMAVAQAQ
** . . : .* ..* :* *..**:.****:******
MMAR_0379|M.marinum_M HAVKDMASLLEDKGVAVKYAIHPVAGRMPGHMNVLLAEAEVDYDAMKDMD
MUL_4733|M.ulcerans_Agy99 HAVKDMASLLEDKGVAVKYAIHPVAGRMPGHMNVLLAEAEVDYDAMKDMD
MAV_5140|M.avium_104 HAVKDMAALLEEKGVPVKYAIHPVAGRMPGHMNVLLAEAEVDYDAMKDMD
MLBr_02634|M.leprae_Br4923 HVVKDMASLLENKGVPVSYAIHPVAGRMPGHMNVLLAEAEVDYDAMKDMD
Mb0162|M.bovis_AF2122/97 HAVKDLATLLEDRGVPVKYAIHPVAGRMPGHMNVLLAEAEVDYDAMKDMD
Rv0157|M.tuberculosis_H37Rv HAVKDLATLLEDRGVPVKYAIHPVAGRMPGHMNVLLAEAEVDYDAMKDMD
TH_0162|M.thermoresistible__bu HAVKDLSTLLESRGVPVKFGIHPVAGRMPGHMNVLLAEAEVDYDAMKDMD
MAB_4577c|M.abscessus_ATCC_199 HTVKEMADLLESKGVEVKYAIHPVAGRMPGHMNVLLAEADVEYDAMKEMD
Mflv_0727|M.gilvum_PYR-GCK YGVADLTRKLRERGVDVRFGIHPVAGRLPGHMNVLLAEAKVPYDIVLEMD
Mvan_0119|M.vanbaalenii_PYR-1 YGVADLTRKLRDRGVDVRFGIHPVAGRLPGHMNVLLAEAKVPYDIVLEMD
MSMEG_0109|M.smegmatis_MC2_155 YGVADLTRKLRERGVNVRFGIHPVAGRLPGHMNVLLAEAKVPYDIVLEMD
: * ::: *..:** * :.*******:***********.* ** : :**
MMAR_0379|M.marinum_M DINDEFARTDVAIVIGANDVTNPAARNDASSPIYGMPILNVDKAKSVIVL
MUL_4733|M.ulcerans_Agy99 DINDEFARTDVAIVIGANDVTNPAARNDASSPIYGMPILNVDKAKSVIVL
MAV_5140|M.avium_104 DINDEFARTDVAIVIGANDVTNPAARNEASSPIYGMPILNVDKAKSVIVL
MLBr_02634|M.leprae_Br4923 DINDEFARTDVVIVIGANDVTNPAARNETASPIYGMPILNVDKAKSVIVL
Mb0162|M.bovis_AF2122/97 DINDEFARTDVTIVIGANDVTNPAARNETSSPIYGMPILNVDKSRSVIVL
Rv0157|M.tuberculosis_H37Rv DINDEFARTDVTIVIGANDVTNPAARNETSSPIYGMPILNVDKSRSVIVL
TH_0162|M.thermoresistible__bu EINDEFARTDVTVVIGANDVTNPAARNDPSSPIHGMPILNVDQSKSVIVL
MAB_4577c|M.abscessus_ATCC_199 DINGEFNRTDVTIVIGANDVTNPAARNDPSSPIHGMPILNVDESRSVIVL
Mflv_0727|M.gilvum_PYR-GCK EINDDFDGTSVVLVIGANDTVNPAASEDPSSPIAGMPVLTVWNADNVIVF
Mvan_0119|M.vanbaalenii_PYR-1 EINDDFDGTSVVLVIGANDTVNPAASEDPGSPIAGMPVLTVWNADNVIVF
MSMEG_0109|M.smegmatis_MC2_155 EINDDFDGTDVVLVIGANDTVNPAAAEDPGSPIAGMPVLQVWNADNVIVF
:**.:* *.*.:******..**** ::..*** ***:* * :: .***:
MMAR_0379|M.marinum_M KRSMNSGFAGIDNPLFYGEGTTMLFGDAKKSVTEVAEELKAL---
MUL_4733|M.ulcerans_Agy99 KRSMNSGFAGIDNPLFYGEGTTMLFGDAKKSVTEVAEELKAL---
MAV_5140|M.avium_104 KRSMNSGFAGIDNPLFYAEGTTMLFGDAKKSVTEVAEELKAL---
MLBr_02634|M.leprae_Br4923 KRSMNSGFAGIDNRLFYADGTTMLFGDAKQSVTEVTEELKVL---
Mb0162|M.bovis_AF2122/97 KRSMNSGFAGIDNPLFYADGTTMLFGDAKKSVTEVSEELKAL---
Rv0157|M.tuberculosis_H37Rv KRSMNSGFAGIDNPLFYADGTTMLFGDAKKSVTEVSEELKAL---
TH_0162|M.thermoresistible__bu KRSMSSGFAGIDNPLFYAEGTTMLFGDAKKSVTEVIEELKAL---
MAB_4577c|M.abscessus_ATCC_199 KRSMSSGYAGIENPLFFQPQTSMLFGDAKKSVSAVIEELKAL---
Mflv_0727|M.gilvum_PYR-GCK KRSMASGYAGVQNPLFFRENTQMLFGDAKDRVDAINAALSETAKV
Mvan_0119|M.vanbaalenii_PYR-1 KRSMASGYAGVQNPLFFRENTQMLFGDAKDRVDAINAALSVAEHV
MSMEG_0109|M.smegmatis_MC2_155 KRSMASGYAGVQNPLFFRDNTQMLFGDARDRVNDILAALPAPERV
**** **:**::* **: * ******:. * : *