For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NSPAHALTKPRRRTPRRAVRLARRALVGLTAAAILAGTGMGWVTYHGALDGITTSNALAGESGSVGDTEN ILIMGLDSRLDQHGNPLPEDVYQALHAGDETVGGYNANVLIVVHLPGDGGPATAFSIPRDDYVDLAGCPS GICKGKVKQAYGFAYQAEMEATESEEEDPATREQKAREAGRKAEIATVRNLLGIPIDHFVEVTLGAFFEI AKAVAPITVCLNDDTSDPYSGADFRKGVQQIDAAQAMAFVRQRRDINDENFTDLDRTRRQQAFLAALVSA LRQSGALSSPSKLRGLLDVTKQNVALDAGFDLASFTKRASALTDAPPTLYTLPIKEFGQNAYGEDINIID VPTIRKIVHDLVSKDDPATTTATPDKPELNGHGATLDVINGSTYTGLAAKLQTTFAADNFTEGQIGDAEA LEPATTIDYGPGADAAAHSLATELGVTATPSDWVLADTVRLTIGTDFPGTDYLGADMFTSTTSGASESES YGPSETAELTPITTVAATATGTYTPAPTDLSQMTASGIPCVK*
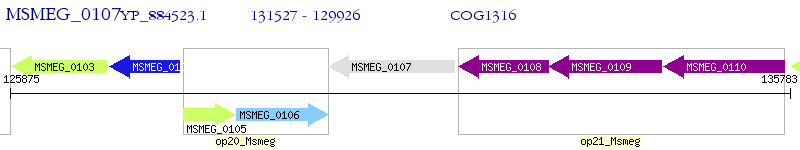
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0107 | - | - | 100% (533) | cell envelope-related function transcriptional attenuator |
| M. smegmatis MC2 155 | MSMEG_1824 | - | 3e-62 | 33.54% (486) | transcriptional regulator, LytR family protein |
| M. smegmatis MC2 155 | MSMEG_6421 | - | 1e-26 | 28.82% (347) | transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3514 | cpsA | 3e-85 | 38.39% (534) | hypothetical protein Mb3514 |
| M. gilvum PYR-GCK | Mflv_4737 | - | 5e-63 | 34.21% (497) | cell envelope-related transcriptional attenuator |
| M. tuberculosis H37Rv | Rv3484 | cpsA | 3e-85 | 38.39% (534) | hypothetical protein Rv3484 |
| M. leprae Br4923 | MLBr_02247 | cpsA | 3e-77 | 36.45% (546) | hypothetical protein MLBr_02247 |
| M. abscessus ATCC 19977 | MAB_3460c | - | 1e-152 | 53.98% (515) | hypothetical protein MAB_3460c |
| M. marinum M | MMAR_4966 | cpsA | 5e-86 | 37.96% (540) | membrane protein CpsA |
| M. avium 104 | MAV_5148 | - | 1e-151 | 54.75% (526) | cell envelope-related function transcriptional attenuator |
| M. thermoresistible (build 8) | TH_3471 | - | 3e-57 | 32.51% (486) | PUTATIVE cell envelope-related transcriptional attenuator |
| M. ulcerans Agy99 | MUL_4040 | cpsA | 3e-84 | 38.18% (516) | membrane protein CpsA |
| M. vanbaalenii PYR-1 | Mvan_1726 | - | 2e-62 | 32.75% (516) | cell envelope-related transcriptional attenuator |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3460c|M.abscessus_ATCC_199 --------------------------MVVGAVAAFVMAITGFGWAGYNTA
MAV_5148|M.avium_104 --------MGSAGTGHPRRWVLLKGRLVASVASAFVMAVTGVGWTGYHTA
MSMEG_0107|M.smegmatis_MC2_155 -MNSPAHALTKPRRRTPRRAVRLARRALVGLTAAAILAGTGMGWVTYHGA
MMAR_4966|M.marinum_M MGRT---KARHRAVRKPSPIRRRVSRTLMTLVSVVALGMTGAGYWVAHGA
MUL_4040|M.ulcerans_Agy99 -----------------------MSRILMTLVSVVALGMTGAGYWVAHGA
Mb3514|M.bovis_AF2122/97 MARSEGNRPRHRAVPQPSRIRKRLSRGVMTLVSVVALLMTGAGYWVAHGA
Rv3484|M.tuberculosis_H37Rv MARSEGNRPRHRAVPQPSRIRKRLSRGVMTLVSVVALLMTGAGYWVAHGA
MLBr_02247|M.leprae_Br4923 MARSHNPRHRHRAIHKPSRIRIALTRSFMTLTSFVAVLLTGAGYWVAHRA
Mflv_4737|M.gilvum_PYR-GCK ------------------MPAAHMLRAVSVALAVAIVIGTGVAWGKIRSF
Mvan_1726|M.vanbaalenii_PYR-1 ------------------MPAAHLVRAISVAMAVVVVLGTGIAWGKIRSF
TH_3471|M.thermoresistible__bu -------------------VPARLIRALAVATAVVIVLSTGVAWSRIRAF
. : : ** .: .
MAB_3460c|M.abscessus_ATCC_199 VGQIITSHVLPGV-LTPVGQDQNILLMGLDSRLDQNGQPLPQDMYDALHA
MAV_5148|M.avium_104 LGRIIISHALPNG-AMSLGDNQNILLMGLDSRLDQNGRPLPPEIYDALHA
MSMEG_0107|M.smegmatis_MC2_155 LDGITTSNALAGE-SGSVGDTENILIMGLDSRLDQHGNPLPEDVYQALHA
MMAR_4966|M.marinum_M LTGITISQALTAEDPRSTGNNMNILLIGLDSRKDQEGNDLPWSILKHLHA
MUL_4040|M.ulcerans_Agy99 LTGITISQALTAEDPRSTGNNMNILLIGLDSRKDQEGNDLPWSILKHLHA
Mb3514|M.bovis_AF2122/97 LGGITISQALTPEDPRSSGNNMNILLIGLDSRKDQEGNDLPWSVLKQLHA
Rv3484|M.tuberculosis_H37Rv LGGITISQALTPEDPRSSGNNMNILLIGLDSRKDQEGNDLPWSVLKQLHA
MLBr_02247|M.leprae_Br4923 LEDITVSQALTTDDPRSTDNNINILLIGLDSRKDQQGNDLPWSILKQLHA
Mflv_4737|M.gilvum_PYR-GCK EAGINHINPIALGGEGEDG-AIDILLVGSDSRTDAHGNPLSDQERAMLRA
Mvan_1726|M.vanbaalenii_PYR-1 ESGINHINPVALGEGGEDG-AIDILLVGSDSRTDAHGNPLSDEERAMLRA
TH_3471|M.thermoresistible__bu EAGISQISSPLLGQGGEDG-AVDILLVGMDSRTDAHGNPLSAEELAALRA
* . :**::* *** * .*. *. . *:*
MAB_3460c|M.abscessus_ATCC_199 GDETSGGYNANVLIVVHIPGNGGPISAVSIPRDDYVNLVGCPGSVCKGKV
MAV_5148|M.avium_104 GDESSGGYNANVLIVVHLSDGDGPVTAVSIPRDDYAELPGCPGSVCAGKI
MSMEG_0107|M.smegmatis_MC2_155 GDETVGGYNANVLIVVHLPGDGGPATAFSIPRDDYVDLAGCPSGICKGKV
MMAR_4966|M.marinum_M GDSDQGGYNTNTLILVHVS-ADGKVVAFSIPRDDWVPFTGVPG-YNHIKI
MUL_4040|M.ulcerans_Agy99 GDSDQGGYNTNTLILVHVS-ADGKVVAFSIPRDDWVPFTGVPG-YNHIKI
Mb3514|M.bovis_AF2122/97 GDSDDGGYNTNTLILVHVG-ADGKVVAFSIPRDDWVPFTGVPG-YNHIKI
Rv3484|M.tuberculosis_H37Rv GDSDDGGYNTNTLILVHVG-ADGKVVAFSIPRDDWVPFTGVPG-YNHIKI
MLBr_02247|M.leprae_Br4923 GDSNDGGYNTNTLILVHVG-ADNKAVAFSIPRDDWVPFSDVPG-YNHIKI
Mflv_4737|M.gilvum_PYR-GCK GDEVST--NTDTIILIRIPNNGESATAISIPRDSYVEAPGIGK----MKI
Mvan_1726|M.vanbaalenii_PYR-1 GDEVST--NTDTIILIRIPNNGQSATAISIPRDSYVESPGIGK----LKI
TH_3471|M.thermoresistible__bu GNDVAT--NTDTIVLVRIPNNGRSATAISIPRDSYVEAPGRGK----MKI
*:. *::.:::::: . *.*****.:. . *:
MAB_3460c|M.abscessus_ATCC_199 KQAYGFAYQQALDAQANGSADAAGAKDLTAREQAAREAGRKAQIDTVSEF
MAV_5148|M.avium_104 KQAYGLAYQQSLNAQAAGNS-GAGATDLAAREQTAREAGRKAEIGAVTDF
MSMEG_0107|M.smegmatis_MC2_155 KQAYGFAYQAEMEATESEEE------DPATREQKAREAGRKAEIATVRNL
MMAR_4966|M.marinum_M KEAYGLTKQYVAQELIDQGV-----TSQRELETRGREAGRAATLRAVRAL
MUL_4040|M.ulcerans_Agy99 KEAYGLTKQYVAQELIDQGV-----TSQRELETRGREAGRAATLRAVRAL
Mb3514|M.bovis_AF2122/97 KEAYGLTKQYVAEQLANQGV-----SDRKELETRGREAARAATLRAVRSL
Rv3484|M.tuberculosis_H37Rv KEAYGLTKQYVAEQLANQGV-----SDRKELETRGREAARAATLRAVRSL
MLBr_02247|M.leprae_Br4923 KEAYGLTKQYVTQKLIDQGV-----TDQKELETQGREAGRAATLRIVRSL
Mflv_4737|M.gilvum_PYR-GCK NGVYGSVHLEEMKELVEQQG-----VDPAEAEPEAQAAGREELIKTVANL
Mvan_1726|M.vanbaalenii_PYR-1 NGVYGSVHLEKMKELVEEQG-----VDPAVAEPEAQAAGREQLIKTVANL
TH_3471|M.thermoresistible__bu NGVYGEVKFETMKQLVEVQG-----MDPEVAEPLAVQDARGALIETVAEL
: .** . . . * . .* : * :
MAB_3460c|M.abscessus_ATCC_199 LGVQIDHFVEVTLAAFFQIAKTVQPIAVCLNHDTIDEFSGANFHEGVQQI
MAV_5148|M.avium_104 LGISVDHFVEVTLGAFFQIAKAVEPITVCLAGDTRDSYSGADFHQGVQRI
MSMEG_0107|M.smegmatis_MC2_155 LGIPIDHFVEVTLGAFFEIAKAVAPITVCLNDDTSDPYSGADFRKGVQQI
MMAR_4966|M.marinum_M TGVPIDYFAEVNLAGFYDLAQALGGVEVCLNHAVDDSYSGADFPAGRQRL
MUL_4040|M.ulcerans_Agy99 TGVPIDYFAEVNLAGFYDLAQALGGVEVCLNHAVDDSYSGADFPAGRQRL
Mb3514|M.bovis_AF2122/97 TGVPIDYFAEINLAGFYDLAQTLGGVDVCLNHAVYDSYSGADFPAGRQRL
Rv3484|M.tuberculosis_H37Rv TGVPIDYFAEINLAGFYDLAQTLGGVDVCLNHAVYDSYSGADFPAGRQRL
MLBr_02247|M.leprae_Br4923 TGVSIDYFAEVNLAGFYDLTHTLGGVEVCLNHAVYDSYSGADFPAGRQRL
Mflv_4737|M.gilvum_PYR-GCK TGVTVDHYAEIGLLGFALITDALGGVDVCLNAPVYEPLSGADFPAGWQKL
Mvan_1726|M.vanbaalenii_PYR-1 TGVTVDHYAEIGLLGFALITDALGGVNVCLNAPVYEPLSGADFPAGWQKL
TH_3471|M.thermoresistible__bu TGVTVDHYAEIGLLGFALITDALGGVEVCLKNPVYEPLSGADFPAGWQRL
*: :*::.*: * .* ::.:: : *** . : ***:* * *::
MAB_3460c|M.abscessus_ATCC_199 DASQAMAFVRQRRDENDGSFTDMDRTRRQQAFLVSLLAAVRQGGAMSNPA
MAV_5148|M.avium_104 DAAEAMAFVRQRRDENDGSFTDFDRTRRQQAFLVSLVEAARSGGALSSVS
MSMEG_0107|M.smegmatis_MC2_155 DAAQAMAFVRQRRDINDENFTDLDRTRRQQAFLAALVSALRQSGALSSPS
MMAR_4966|M.marinum_M DAAQALAFVRQRHGLDNG---DLDRTHRQQAFLSSVMRDLQDSGTFTNLD
MUL_4040|M.ulcerans_Agy99 DAAQALAFVRQRHGLDNG---DLDRTHRQQVFLSSVMRDLQDSGTFTNLD
Mb3514|M.bovis_AF2122/97 NAAQALAFVRQRHGLDNG---DLDRTHRQQAFLSSVMRELQDSGTFTNLD
Rv3484|M.tuberculosis_H37Rv NAAQALAFVRQRHGLDNG---DLDRTHRQQAFLSSVMRELQDSGTFTNLD
MLBr_02247|M.leprae_Br4923 DASEALAFVRQRHGLDNG---DLDRTHRQQAFISSVMRELGDTGTFTNLD
Mflv_4737|M.gilvum_PYR-GCK NGTQALSFVRQRHDLPRG---DLDRVARQQAAMASMAHEVISSKTLSSPS
Mvan_1726|M.vanbaalenii_PYR-1 NGTQALSFVRQRHDLPRG---DLDRVARQQAVMASMAHEVISSKTLSSPT
TH_3471|M.thermoresistible__bu DGAQALSFVRQRQDLPRG---DLDRVTRQQTVMASLAREVISSKTLSSPA
:.::*::*****:. *:**. ***. : :: . :::.
MAB_3460c|M.abscessus_ATCC_199 AIRNILNVAHENVAIDSGLNIVDFAARASELTK----RPTSFYTLPISDF
MAV_5148|M.avium_104 GLRKLLQVARDNVAVDAGLDLMQFAARASQLAG----RPLSLYTLPISGF
MSMEG_0107|M.smegmatis_MC2_155 KLRGLLDVTKQNVALDAGFDLASFTKRASALTD----APPTLYTLPIKEF
MMAR_4966|M.marinum_M RLDSLMAVARKDLVLSGGWD-EDLIRRMGELAG----ADIEFRTLPVVRY
MUL_4040|M.ulcerans_Agy99 RLDSLMAVARKDLVLSGGWD-EDLVRRMGELAG----ADIEFRTLPVVRY
Mb3514|M.bovis_AF2122/97 RLDNLMAVARKDVVLSAGWD-EDLFRRMGDLAG----GNVEFRTLPVVRY
Rv3484|M.tuberculosis_H37Rv RLDNLMAVARKDVVLSAGWD-EDLFRRMGDLAG----GNVEFRTLPVVRY
MLBr_02247|M.leprae_Br4923 KLRSLMAVARKDVVLSAGWN-EDLIQRLGTLANDVGENRTQFRTLPVVRY
Mflv_4737|M.gilvum_PYR-GCK TLSALESAVQRSVVLSEGWDIMDFVEQLQKLAA----GNVAFATIPILEE
Mvan_1726|M.vanbaalenii_PYR-1 TLNNLEDAVQRSVVLSDGWNIMDFVEQLQKLAG----GNIAFATIPIVEE
TH_3471|M.thermoresistible__bu TLDRLQRAIQRSVVLSDGWDVMEFVEQLQGLAA----GNIAFATIPIVRE
: : . : .:.:. * : .: : *: : *:*:
MAB_3460c|M.abscessus_ATCC_199 GAD--SNGSDVNIVDLPTIRQIVHDRFSSDPVPEAPSAAAASAPPAL-ST
MAV_5148|M.avium_104 GKD--PNGSDINLVDLPAIRRIVNERFMSDAPAALDAAVPGSQPPAAPAE
MSMEG_0107|M.smegmatis_MC2_155 GQN--AYGEDINIIDVPTIRKIVHDLVSKDDPATTTATP---DKPELNGH
MMAR_4966|M.marinum_M DN---IDGQDVNIVDPAAIKAEVAAAIGTTSTAATATTT-----PAKPSP
MUL_4040|M.ulcerans_Agy99 DN---IDGQDVNIVDPAAIKAEVAAAIGSTSTAATATTT-----PAKPSP
Mb3514|M.bovis_AF2122/97 DN---IDGQDVNIIDPTAIRAEVAAAFG-SAPPTSQTAA-----AAKPNP
Rv3484|M.tuberculosis_H37Rv DN---IDGQDVNIIDPTAIRAEVAAAFG-SAPPTSQTAA-----AAKPNP
MLBr_02247|M.leprae_Br4923 DN---INGQAVNIIDPVAIKAEVASTIG---TASPATAT-----PSKPNP
Mflv_4737|M.gilvum_PYR-GCK AGWSDDGMQSVVRVDPKEVKDWVTSLLNDQDQGKTEELA-------YSND
Mvan_1726|M.vanbaalenii_PYR-1 AGWSDDGMQSVVRVDPAAVKDWVTGLLHDQDEGKTEELT-------YSND
TH_3471|M.thermoresistible__bu DGWSDDGMQSVVRVDPQQVRDWVSSLLEEQDEGKTEEIA-------YSRD
. : :* :: * .
MAB_3460c|M.abscessus_ATCC_199 PVVLNVVNATSRDGLAAAVEDAFTARGFTRGRASTAETQSTES-SIMYGE
MAV_5148|M.avium_104 PAVLDVVNATTHDGLAAAVEEALAGRGFTRGSATTAPTQAEDS-SIEYGP
MSMEG_0107|M.smegmatis_MC2_155 GATLDVINGSTYTGLAAKLQTTFAADNFTEGQIGDAEALEPAT-TIDYGP
MMAR_4966|M.marinum_M STVVDVVNAGSISGLASQVSNALRKRGYTTDQVRDRKSGDPAATTIKYGP
MUL_4040|M.ulcerans_Agy99 STVVDVVNAGSISGLASQVSNALRKRGYTTDQVRDRKSGDPAATTIKYGP
Mb3514|M.bovis_AF2122/97 STVVDVVNAGSISGLASQVSGALLKRGYTAGQVRDRESGDPFTTAIEYGA
Rv3484|M.tuberculosis_H37Rv STVVDVVNAGSISGLASQVSGALLKRGYTAGQVRDRESGDPFTTAIEYGA
MLBr_02247|M.leprae_Br4923 STVVDVINAGRLTGLASEVFQTLKKHGYTAGQVRDRSSGEPTTTSIQYGA
Mflv_4737|M.gilvum_PYR-GCK KTTVEVYNDSDVNGLAAAVAHVLTGKGFTQGNVGNNEGPKVTGSQVRAAK
Mvan_1726|M.vanbaalenii_PYR-1 KTTVEVVNDTDINGLAAAVSHVLSGKGFTKGNVGNNDGAKVTASQVQAAK
TH_3471|M.thermoresistible__bu KTTVEVLNGTDINGLAAAVSHVLTSNGFSPGPVGNAEDGPADRTEVRVAK
..::* * ***: : .: .:: . : .
MAB_3460c|M.abscessus_ATCC_199 DAQEGAQALADQLHLPMT--ESGAVAPGTVQLTVGSQFPVDNYIAHHSGG
MAV_5148|M.avium_104 GAEAAARLLADQLHLPAT--AQSAVAPGTVRLTVGAAFPAADYLAHP---
MSMEG_0107|M.smegmatis_MC2_155 GADAAAHSLATELGVTAT--PSDWVLADTVRLTIGTDFPGTDYLGADMFT
MMAR_4966|M.marinum_M GAQADAMNLGTLLGIDAPKQPDASIEPGHIQVTVDTNFSMPAPDDSDDDT
MUL_4040|M.ulcerans_Agy99 GAQADAMNLGTLLGIDAPKQPDASIEPGHIQVTVDTNFSMPAPDDSDDDT
Mb3514|M.bovis_AF2122/97 GAETDAQNVADLLGIDAPNHPDPAVAPGHIRVTVDTNFSLPAPDEATAAA
Rv3484|M.tuberculosis_H37Rv GAETDAQNVADLLGIDAPNHPDPAVAPGHIRVTVDTNFSLPAPDEATAAA
MLBr_02247|M.leprae_Br4923 GADTDAQNVATLLSIDAPNQADPKLASGHIRVIVDTNYSLPADEDSTMDE
Mflv_4737|M.gilvum_PYR-GCK VDDMGAQAVALDLG-GLEVVEDSSVPPDTVRVVLADDYTGPGAGLDGSDV
Mvan_1726|M.vanbaalenii_PYR-1 TDDLGAQAVAQELG-GLPVVENAQVPQGSVRVVLAADYTGPGAGLDGSDV
TH_3471|M.thermoresistible__bu QDDRGAREVADQLG-DLPIVEDRTVAPGFVRVVLADDYTGPGSGLDGTDP
: * :. * . : . ::: : :.
MAB_3460c|M.abscessus_ATCC_199 AKGGSSGEAD--------PADKMTAVAATGTGVQAPAPTDLSQLTANGIP
MAV_5148|M.avium_104 ---GASG------------SGQVSTVSATGTGDHAPAPTDLSQMTATSVP
MSMEG_0107|M.smegmatis_MC2_155 STTSGASESESYGPSETAELTPITTVAATATGTYTPAPTDLSQMTASGIP
MMAR_4966|M.marinum_M TTYPKPN---------------AYYNGGDD-SSSYPAPDHGQPIDGGGVP
MUL_4040|M.ulcerans_Agy99 TVYPKPN---------------AYYNGGDD-SSSYPAPDHGQPIDGGGVP
Mb3514|M.bovis_AF2122/97 TSTETST---------------YPLYGGG--TTTDPTPDQGAPIDGGGVP
Rv3484|M.tuberculosis_H37Rv TSTETST---------------YPLYGGG--TTTDPTPDQGAPIDGGGVP
MLBr_02247|M.leprae_Br4923 TITSQKV---------------GTYPPYSYNTSPYPTPDQGKPIDGGGIP
Mflv_4737|M.gilvum_PYR-GCK TLMGADPST---------------SGVVSDTGAAVPAPSPIITAGSDDPK
Mvan_1726|M.vanbaalenii_PYR-1 TLMGADPST---------------SGVVTDTGATVPAPPPILTAGSNDPE
TH_3471|M.thermoresistible__bu TVTTLQQS----------------SATVGQTVEAAPPPPQIITAGADDPK
*.* . .
MAB_3460c|M.abscessus_ATCC_199 CVK
MAV_5148|M.avium_104 CVK
MSMEG_0107|M.smegmatis_MC2_155 CVK
MMAR_4966|M.marinum_M CVN
MUL_4040|M.ulcerans_Agy99 CVN
Mb3514|M.bovis_AF2122/97 CVN
Rv3484|M.tuberculosis_H37Rv CVN
MLBr_02247|M.leprae_Br4923 CVN
Mflv_4737|M.gilvum_PYR-GCK CVN
Mvan_1726|M.vanbaalenii_PYR-1 CVN
TH_3471|M.thermoresistible__bu CVN
**: