For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PAAHLVRAISVAMAVVVVLGTGIAWGKIRSFESGINHINPVALGEGGEDGAIDILLVGSDSRTDAHGNPL SDEERAMLRAGDEVSTNTDTIILIRIPNNGQSATAISIPRDSYVESPGIGKLKINGVYGSVHLEKMKELV EEQGVDPAVAEPEAQAAGREQLIKTVANLTGVTVDHYAEIGLLGFALITDALGGVNVCLNAPVYEPLSGA DFPAGWQKLNGTQALSFVRQRHDLPRGDLDRVARQQAVMASMAHEVISSKTLSSPTTLNNLEDAVQRSVV LSDGWNIMDFVEQLQKLAGGNIAFATIPIVEEAGWSDDGMQSVVRVDPAAVKDWVTGLLHDQDEGKTEEL TYSNDKTTVEVVNDTDINGLAAAVSHVLSGKGFTKGNVGNNDGAKVTASQVQAAKTDDLGAQAVAQELGG LPVVENAQVPQGSVRVVLAADYTGPGAGLDGSDVTLMGADPSTSGVVTDTGATVPAPPPILTAGSNDPEC VN*
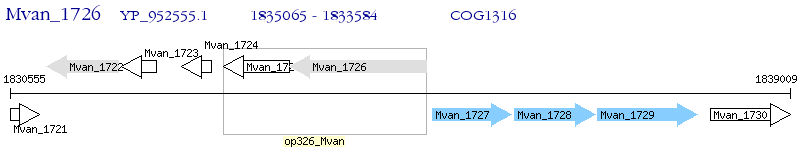
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1726 | - | - | 100% (493) | cell envelope-related transcriptional attenuator |
| M. vanbaalenii PYR-1 | Mvan_5665 | - | 1e-36 | 35.52% (290) | cell envelope-related transcriptional attenuator |
| M. vanbaalenii PYR-1 | Mvan_5087 | - | 1e-25 | 24.23% (487) | cell envelope-related transcriptional attenuator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3295 | - | 0.0 | 72.02% (486) | hypothetical protein Mb3295 |
| M. gilvum PYR-GCK | Mflv_4737 | - | 0.0 | 86.82% (493) | cell envelope-related transcriptional attenuator |
| M. tuberculosis H37Rv | Rv3267 | - | 0.0 | 72.02% (486) | hypothetical protein Rv3267 |
| M. leprae Br4923 | MLBr_00750 | - | 0.0 | 64.85% (495) | hypothetical protein MLBr_00750 |
| M. abscessus ATCC 19977 | MAB_3614 | - | 1e-159 | 60.81% (495) | hypothetical protein MAB_3614 |
| M. marinum M | MMAR_1274 | - | 0.0 | 69.78% (493) | hypothetical protein MMAR_1274 |
| M. avium 104 | MAV_4232 | - | 0.0 | 69.42% (484) | cell envelope-related function transcriptional attenuator |
| M. smegmatis MC2 155 | MSMEG_1824 | - | 0.0 | 75.30% (494) | transcriptional regulator, LytR family protein |
| M. thermoresistible (build 8) | TH_3471 | - | 0.0 | 70.82% (490) | PUTATIVE cell envelope-related transcriptional attenuator |
| M. ulcerans Agy99 | MUL_2613 | - | 0.0 | 69.57% (493) | hypothetical protein MUL_2613 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1726|M.vanbaalenii_PYR-1 -----------------------------MPAAHLVR------AISVAMA
Mflv_4737|M.gilvum_PYR-GCK -----------------------------MPAAHMLR------AVSVALA
MSMEG_1824|M.smegmatis_MC2_155 ----------------------------------MIR------SIAVAAA
TH_3471|M.thermoresistible__bu ------------------------------VPARLIR------ALAVATA
MMAR_1274|M.marinum_M -----------------------------MPVQRVVR------AVATALA
MUL_2613|M.ulcerans_Agy99 ----------------------------MMPVQRVVR------AVATALA
Mb3295|M.bovis_AF2122/97 ----------------------------MMSAQRVVRTVRTARAISTALA
Rv3267|M.tuberculosis_H37Rv ----------------------------MMSAQRVVRTVRTARAISTALA
MLBr_00750|M.leprae_Br4923 -----------------------------MPVQRVVR------AVVTALT
MAV_4232|M.avium_104 -------------------------------------------MIATALT
MAB_3614|M.abscessus_ATCC_1997 MPGEGPAGEEWTDIVTDPHARRASGSSQAEPPRPLWR------GIATLAA
: . :
Mvan_1726|M.vanbaalenii_PYR-1 VVVVLGTGIAWGKIRSFESGINHINPV-ALGEGG--EDGAIDILLVGSDS
Mflv_4737|M.gilvum_PYR-GCK VAIVIGTGVAWGKIRSFEAGINHINPI-ALGGEG--EDGAIDILLVGSDS
MSMEG_1824|M.smegmatis_MC2_155 SAVVLGTGVAWTQVRSFESGINHISSS-ALGEGG--EDGAIDILLVGIDS
TH_3471|M.thermoresistible__bu VVIVLSTGVAWSRIRAFEAGISQISSP-LLGQGG--EDGAVDILLVGMDS
MMAR_1274|M.marinum_M VVVVLGTGLAWTNVRSFEGGIFHMSAP-SLGHGA--DDGAIDILLVGLDS
MUL_2613|M.ulcerans_Agy99 VVVVLGTGLAWTNVRSFEGGIFHMSAP-SLGHGA--DDGAIDILLVGLDS
Mb3295|M.bovis_AF2122/97 VAIVLGTGVAWSSVRSFEDGIFHMSAP-SLGHGG--DDGAIDILLVGLDS
Rv3267|M.tuberculosis_H37Rv VAIVLGTGVAWSSVRSFEDGIFHMSAP-SLGHGG--DDGAIDILLVGLDS
MLBr_00750|M.leprae_Br4923 LAIVLGTGITWTSFRSFEDGIFHMSAP-SLGKGG--NDGAIDILLVGLDS
MAV_4232|M.avium_104 LAVVLGTGIAWSNVRSFEDGIFHMSAP-SLGKGG--DDGAIDILLVGLDS
MAB_3614|M.abscessus_ATCC_1997 VAVMVITGAAWGKIGNIDPNIARFDLPGLFGNAGKPDDGAIDILMVGVDS
.::: ** :* . :: .* ::. :* . :***:***:** **
Mvan_1726|M.vanbaalenii_PYR-1 RTDAHGNPLSDEERAMLRAGDEVSTNTDTIILIRIPNNGQSATAISIPRD
Mflv_4737|M.gilvum_PYR-GCK RTDAHGNPLSDQERAMLRAGDEVSTNTDTIILIRIPNNGESATAISIPRD
MSMEG_1824|M.smegmatis_MC2_155 RTDAHGNPLSPEELATLRAGDDVSTNTDTIILVRIPNNGTSATAISIPRD
TH_3471|M.thermoresistible__bu RTDAHGNPLSAEELAALRAGNDVATNTDTIVLVRIPNNGRSATAISIPRD
MMAR_1274|M.marinum_M RTDAHGNPLTPEELATLRAGDEEATNTDTIILVRIPNNGRSATAISIPRD
MUL_2613|M.ulcerans_Agy99 RTDAHGNPLTPEELATLRAGDEEATNTDTIILVRIPNNGRSATAISIPRD
Mb3295|M.bovis_AF2122/97 RTDAHGNPLSAEELATLHAGDEEATNTDTIILIRVPNNGKSATAISIPRD
Rv3267|M.tuberculosis_H37Rv RTDAHGNPLSAEELATLHAGDEEATNTDTIILIRVPNNGKSATAISIPRD
MLBr_00750|M.leprae_Br4923 RTDAYGNPLSAEELATLRAGDEEATNTDTIILIRIPNNGKSATAISIPRD
MAV_4232|M.avium_104 RTDAHGNPLSQQELETLRAGDEEATNTDTIILIRIPNNGKSATAISIPRD
MAB_3614|M.abscessus_ATCC_1997 RSDAHGNPLSQEELSMLRAGDETATNTDTIILIRIPNNGKSATAISIPRD
*:**:****: :* *:**:: :******:*:*:**** **********
Mvan_1726|M.vanbaalenii_PYR-1 SYVESPGIGKLKINGVYGSVHLEKMKELVEEQGVDPAVAEPEAQAAGREQ
Mflv_4737|M.gilvum_PYR-GCK SYVEAPGIGKMKINGVYGSVHLEEMKELVEQQGVDPAEAEPEAQAAGREE
MSMEG_1824|M.smegmatis_MC2_155 SYVEAPGWGKMKINGVFGDVKLDRMKQLVEVEGEDPAVAEPEATEAGREK
TH_3471|M.thermoresistible__bu SYVEAPGRGKMKINGVYGEVKFETMKQLVEVQGMDPEVAEPLAVQDARGA
MMAR_1274|M.marinum_M SYVQAPGLGKTKINGVYGQTRETKRAGLVQ-AGASAAEAAAAGTEAGREA
MUL_2613|M.ulcerans_Agy99 SYVQAPGLGKTKINGVYGQTRETKRAGLVQ-AGASAAEAAAAGTEAGREA
Mb3295|M.bovis_AF2122/97 SYVAAPGLGKTKINGVYGQTRETKRAGLVQ-AGASPTEAAAAGTEAGREA
Rv3267|M.tuberculosis_H37Rv SYVAAPGLGKTKINGVYGQTRETKRAGLVQ-AGASPTEAAAAGTEAGREA
MLBr_00750|M.leprae_Br4923 SYVQAPGLGKAKINGVYGQTREAKRASLVR-TGDSAEDAAAQGTEAGREA
MAV_4232|M.avium_104 SYVAAPGLGKTKINGVYGQTREAKRASLVK-AGDSAEDAAAQGTEAGREA
MAB_3614|M.abscessus_ATCC_1997 SYVTIPDGSKGKINGVYGEAKENDRQKRVE-SGETLEAAERESVDAGRSA
*** *. .* *****:*..: *. * * . .*
Mvan_1726|M.vanbaalenii_PYR-1 LIKTVANLTGVTVDHYAEIGLLGFALITDALGGVNVCLNAPVYEPLSGAD
Mflv_4737|M.gilvum_PYR-GCK LIKTVANLTGVTVDHYAEIGLLGFALITDALGGVDVCLNAPVYEPLSGAD
MSMEG_1824|M.smegmatis_MC2_155 LIETVADLTGVTVDHYAEIGLLGFALITDALGGVNVCLNEPVYEPLSGAD
TH_3471|M.thermoresistible__bu LIETVAELTGVTVDHYAEIGLLGFALITDALGGVEVCLKNPVYEPLSGAD
MMAR_1274|M.marinum_M LIKTVADLTGVTVDHYAEIGLLGFALITDALGGVDVCLNGPVYEPLSGAD
MUL_2613|M.ulcerans_Agy99 LIKTVADLTGVTVDHYAEIGLLGFALITDALGGVDVCLNGPVYEPLSGAD
Mb3295|M.bovis_AF2122/97 LIKTVADLTGVTVDHYAEIGLLGFALIADALGGVDVCLKEPVYEPLSGAD
Rv3267|M.tuberculosis_H37Rv LIKTVADLTGVTVDHYAEIGLLGFALIADALGGVDVCLKEPVYEPLSGAD
MLBr_00750|M.leprae_Br4923 LIKTVADLTGVTVDHYAEIGLLGFALIVDALGGVDVCLKDAVFEPMSGAD
MAV_4232|M.avium_104 LIKTVADLTGVTVDHYAEIGLLGFALITDALGGVDVCLKEPVFEPLSGAD
MAB_3614|M.abscessus_ATCC_1997 LIQTVGRLTGVTVDRYAEVSMLGFVLMTDALGGVNVCLNEAVYEPMSGAD
**:**. *******:***:.:***.*:.******:***: .*:**:****
Mvan_1726|M.vanbaalenii_PYR-1 FPAGWQKLNGTQALSFVRQRHDLPRGDLDRVARQQAVMASMAHEVISSKT
Mflv_4737|M.gilvum_PYR-GCK FPAGWQKLNGTQALSFVRQRHDLPRGDLDRVARQQAAMASMAHEVISSKT
MSMEG_1824|M.smegmatis_MC2_155 FPAGWQKLNGPQALSFVRQRHGLPRGDLDRVTRQQAFMASLAHEAISSKT
TH_3471|M.thermoresistible__bu FPAGWQRLDGAQALSFVRQRQDLPRGDLDRVTRQQTVMASLAREVISSKT
MMAR_1274|M.marinum_M FPAGRQKLTGPQALAFVRQRHELPRGDLDRVVRQQAVMASLAHRVISGKT
MUL_2613|M.ulcerans_Agy99 FPAGRQKLTGPQALAFVRQRHELPRGDLDRVVRQQAVMASLAHRVISGKT
Mb3295|M.bovis_AF2122/97 FPAGRQKLNGPQALSFVRQRHDLPRGDLDRVVRQQAVMAALAHRVISGQT
Rv3267|M.tuberculosis_H37Rv FPAGRQKLNGPQALSFVRQRHDLPRGDLDRVVRQQAVMAALAHRVISGQT
MLBr_00750|M.leprae_Br4923 FPAGRQKLNGPQALSFVRQRHELPRGDLDRVVRQQAVMASLVHRVISGQT
MAV_4232|M.avium_104 FPAGPQRLSGPEALSFVRQRHELPRGDLDRVVRQQVVMASLAHRVISGQT
MAB_3614|M.abscessus_ATCC_1997 FPAGPQTLDGPNALSFVRQRHDLPRGDLDRVVRQQVVMSSLAHSALSGGT
**** * * *.:**:*****: *********.***. *:::.: .:*. *
Mvan_1726|M.vanbaalenii_PYR-1 LSSPTTLNNLEDAVQRSVVLSDGWNIMDFVEQLQKLAGGNIAFATIPIVE
Mflv_4737|M.gilvum_PYR-GCK LSSPSTLSALESAVQRSVVLSEGWDIMDFVEQLQKLAAGNVAFATIPILE
MSMEG_1824|M.smegmatis_MC2_155 LSSPATLGRLQDAVQRSVVISDGWDIMDFVSQLQNLAGGNVAFATIPILR
TH_3471|M.thermoresistible__bu LSSPATLDRLQRAIQRSVVLSDGWDVMEFVEQLQGLAAGNIAFATIPIVR
MMAR_1274|M.marinum_M LSSPTTVRRLEQAVQRSVVLSSGWNIIDFVQQLQQLTGGNVAFATIPVLD
MUL_2613|M.ulcerans_Agy99 LSSPTTVRRLEQAVQRSVVLSSGWNIIDFVQQLQQLTGGNVAFATIPVLD
Mb3295|M.bovis_AF2122/97 LSSPATLKRLEQAVQRSVVLSSGWDIMDFVRQLQKLAGGNVAFATIPVLD
Rv3267|M.tuberculosis_H37Rv LSSPATLKRLEQAVQRSVVLSSGWDIMDFVRQLQKLAGGNVAFATIPVLD
MLBr_00750|M.leprae_Br4923 LSNPSTIKHLEQAIQRSVVISSGWNLMDFLKQLQSLADRKVAFATIPVLD
MAV_4232|M.avium_104 LSSPATVKRLEAAVQRSVVISAGWDVMDFVQQMQKLAGGNVAFATIPVLD
MAB_3614|M.abscessus_ATCC_1997 LTNTTTLGKLRDAITRTVVLSQGWDVMDFIQQLQKLSGGNVAFATIPILR
*:..:*: *. *: *:**:* **::::*: *:* *: ::******::
Mvan_1726|M.vanbaalenii_PYR-1 EAGWSDDGMQSVVRVDPAAVKDWVTGLLHDQDEGKTEELTYSNDKTTVEV
Mflv_4737|M.gilvum_PYR-GCK EAGWSDDGMQSVVRVDPKEVKDWVTSLLNDQDQGKTEELAYSNDKTTVEV
MSMEG_1824|M.smegmatis_MC2_155 EDGWSDDGMQSVVRVDPQQVRDWVTGLLQDQDEGKTEELAYSPAKTTVEV
TH_3471|M.thermoresistible__bu EDGWSDDGMQSVVRVDPQQVRDWVSSLLEEQDEGKTEEIAYSRDKTTVEV
MMAR_1274|M.marinum_M GSGWSDDGMQSVVRVDPHQVADWVGSLLNEQNEGKTAELAYAPAKTTADV
MUL_2613|M.ulcerans_Agy99 GSGWSDDGMQSVVRVDPHQVADWVGSLLNEQNEGKTAELAYAPAKTTADV
Mb3295|M.bovis_AF2122/97 GAGWSDDGMQSVVRVDPRQVQDWVVGLLHEQDQGKTDELAYTPAKTTANV
Rv3267|M.tuberculosis_H37Rv GAGWSDDGMQSVVRVDPRQVQDWVVGLLHEQDQGKTDELAYTPAKTTANV
MLBr_00750|M.leprae_Br4923 ETGWSDDGMQSVVRVDPHQVQNFVSGLLHNQAQDKTDELAYTPAKITANV
MAV_4232|M.avium_104 GAGWSDDGMQSVVRVDPHQVADWVGSLLHDQEQGKTEEIAYTPAKTTASV
MAB_3614|M.abscessus_ATCC_1997 EDGWTEDGTQSVVKVDPDQVRSWVSGLLDQQAQGKTEEATYSKDQTTADV
**::** ****:*** * .:* .**.:* :.** * :*: : *..*
Mvan_1726|M.vanbaalenii_PYR-1 VNDTDINGLAAAVSHVLSGKGFTKGNVGNNDGAKVTASQVQAAKTDDLGA
Mflv_4737|M.gilvum_PYR-GCK YNDSDVNGLAAAVAHVLTGKGFTQGNVGNNEGPKVTGSQVRAAKVDDMGA
MSMEG_1824|M.smegmatis_MC2_155 LNGTDVNGLAASVSQVLTQKGFTPGDTGNHEDGPVAGSQVLAATSDDLGA
TH_3471|M.thermoresistible__bu LNGTDINGLAAAVSHVLTSNGFSPGPVGNAEDGPADRTEVRVAKQDDRGA
MMAR_1274|M.marinum_M VNDTDINGLAAAVSDVLSAKGFTTGSVGNNEAGHVKLSQVRAATTDDLGA
MUL_2613|M.ulcerans_Agy99 VNDTDINGLAAAVSDVLSAKGFTTGSVGNNEAGHVKLSQVRAVTTDDLGA
Mb3295|M.bovis_AF2122/97 VNDTDINGLAAAVSKVLSSKGFTTGSVGNNDGDHVPGSQVRAAKADDLGA
Rv3267|M.tuberculosis_H37Rv VNDTDINGLAAAVSKVLSSKGFTTGSVGNNDGDHVPGSQVRAAKADDLGA
MLBr_00750|M.leprae_Br4923 ANDTDINGLAAAVSDILTTKGFIAGSVANNQGSHVKSSQVRAAKSDDLGA
MAV_4232|M.avium_104 VNDTDINGLAAAVSDVLSAKGFATGTVGNNDGGHVKTSQVRAAKSDDLGA
MAB_3614|M.abscessus_ATCC_1997 VNETQINGLAAAVSELLVGKGFTAGKVGNNESDHVGSSQVQAAQQDDVGA
* :::*****:*:.:* :** * ..* : . ::* .. ** **
Mvan_1726|M.vanbaalenii_PYR-1 QAVAQELGGLPVVENAQVPQGSVRVVLAADYTGPGAGLDGS--DVTLMGA
Mflv_4737|M.gilvum_PYR-GCK QAVALDLGGLEVVEDSSVPPDTVRVVLADDYTGPGAGLDGS--DVTLMGA
MSMEG_1824|M.smegmatis_MC2_155 QEVAKDLGGLPVHEDSSIPAGSVRVVLGDDYTGPGSGLDG---NPALSSV
TH_3471|M.thermoresistible__bu REVADQLGDLPIVEDRTVAPGFVRVVLADDYTGPGSGLDGT--DPTVTTL
MMAR_1274|M.marinum_M QQVAKELGGLPVVADTSIPSGSVRVVLSNDYSGPGSGLGG-----AMAPA
MUL_2613|M.ulcerans_Agy99 QQVAKELGGLPVVADTSIPSGSVRVVLSNDYSGPGSGLGG-----AMAPA
Mb3295|M.bovis_AF2122/97 QQVAKELGGLPVVADASIAPGSVRVVLANDYSGPGSGLGGSDPNGVVSPA
Rv3267|M.tuberculosis_H37Rv QQVAKELGGLPVVADASIAPGSVRVVLANDYSGPGSGLGGSDPNGVVSPA
MLBr_00750|M.leprae_Br4923 QKVSKELGGLPIVVDGSIPAGSVLVILANDYTGPGSGLFG---NGSIASD
MAV_4232|M.avium_104 KEVSKELGGLPVVADSSLAPGAVRVVLANDYSGPGSGLSG---TESIMPA
MAB_3614|M.abscessus_ATCC_1997 KAVAEALG-LPVVEKQGIAEHTVRVVLSKDYQGPGSGRET-----TPAAS
: *: ** * : . :. * *:*. ** ***:*
Mvan_1726|M.vanbaalenii_PYR-1 DPSTSGVVT----DTGATVPAPP--PILTAGSNDPECVN
Mflv_4737|M.gilvum_PYR-GCK DPSTSGVVS----DTGAAVPAPS--PIITAGSDDPKCVN
MSMEG_1824|M.smegmatis_MC2_155 DPASSSASGYADGSSEDETPPPPPSPILTAGANDPQCVN
TH_3471|M.thermoresistible__bu QQSSATVG-----QTVEAAPPPP--QIITAGADDPKCVN
MMAR_1274|M.marinum_M HASN--PGY-----S-DTAPPPS--PILTAGSDQPECIN
MUL_2613|M.ulcerans_Agy99 HASN--PGY-----S-DTAPPPS--PILTAGSDQPECIN
Mb3295|M.bovis_AF2122/97 RAFN--LGS-----ADDTTPPPS--PILTAGSDAPECIN
Rv3267|M.tuberculosis_H37Rv RAFN--LGS-----ADDTTPPPS--PILTAGSDAPECIN
MLBr_00750|M.leprae_Br4923 YASNPGAS------ADPNIPVPS--PILTAGSDQPECIN
MAV_4232|M.avium_104 KVSTAGAVS-----TNDKAPAPS--PILTAGSDKPECIN
MAB_3614|M.abscessus_ATCC_1997 ETGS----------TEEDAPPPPPSPIFTA-ANGPRCVN
. : * *. *:** :: *.*:*