For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPNSYRVVQWTTGNVGKSSLQAIATHPALELVGCYAWSQEKAGRDAGELCGMAPLGITASNDVEALLALK PDCVVYNPMWIDVDELVRILSAGVNVVTTAAFITGHNLGAGRDRILAACRHGGSTIFGSGVSPGFAELLA IVSAMVCNRIDKVTVNEAADTTFYDSPATERPVGFGQPIDHPQLQAMTAQGTAIFGEAVRLVADALGVEL DEVRCVAEHAQTTADLDLGSWTIAAGCVAGVHASWQGVAGGRTIVELNVRWRKGQTLEPDWQIDQDGWVI QVDGTPTVTTKVSFLPPPYFQAETIADFMVLGHIMTAMPTINAIPAVVAAAPGIVTYSDLALTLPRGMVP TDRNSDPKLGASRTP
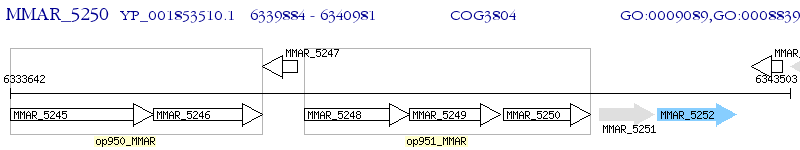
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5250 | - | - | 100% (365) | hypothetical protein MMAR_5250 |
| M. marinum M | MMAR_3741 | - | 3e-74 | 45.10% (337) | hypothetical protein MMAR_3741 |
| M. marinum M | MMAR_1936 | - | 2e-63 | 35.71% (350) | hypothetical protein MMAR_1936 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1088 | - | 5e-45 | 33.62% (354) | hypothetical protein Mb1088 |
| M. gilvum PYR-GCK | Mflv_1277 | - | 1e-147 | 70.42% (355) | dihydrodipicolinate reductase |
| M. tuberculosis H37Rv | Rv1059 | - | 5e-45 | 33.62% (354) | hypothetical protein Rv1059 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0727 | - | 2e-65 | 37.28% (346) | putative dihydrodipicolinate reductase |
| M. avium 104 | MAV_0371 | dapB | 1e-168 | 81.27% (347) | dihydrodipicolinate reductase |
| M. smegmatis MC2 155 | MSMEG_0560 | dapB | 1e-162 | 76.08% (347) | dihydrodipicolinate reductase N-terminus domain-containing |
| M. thermoresistible (build 8) | TH_4249 | - | 1e-150 | 71.71% (350) | PUTATIVE dihydrodipicolinate reductase |
| M. ulcerans Agy99 | MUL_3685 | - | 1e-73 | 44.81% (337) | hypothetical protein MUL_3685 |
| M. vanbaalenii PYR-1 | Mvan_5538 | - | 1e-149 | 70.99% (355) | dihydrodipicolinate reductase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5250|M.marinum_M MPNS--YRVVQWTTGNVGKSSLQAIATHPALELVGCYAWSQEKAGRDAGE
MAV_0371|M.avium_104 MPNT--YRVVQWNTGNVGKSSLKSIVTNPTLELVGCYAWSAEKVGRDAGE
MSMEG_0560|M.smegmatis_MC2_155 MPDTTGYRVVQWTTGNVGKSSVAAIAKNPTLELVGCYAWSGDKVGKDVGE
Mflv_1277|M.gilvum_PYR-GCK MPNNAPHRVVQWTTGNVGKSSVAAITANPTLELVGCYAWSDDKVGRDVGE
Mvan_5538|M.vanbaalenii_PYR-1 MPNTASYRVVQWTTGNVGKSSVAAIAGNPTLELVGCYAWSDDKVGRDAGE
TH_4249|M.thermoresistible__bu VPN-TPYRVVQWTTGNVGKSSVQALAANPLYELVGVYAWSPDKVGRDAGE
MAB_0727|M.abscessus_ATCC_1997 ---MSVIRVFQVATGNVGTEMIRRIASHPDLELAGLHCYSPEKVGRDAGE
MUL_3685|M.ulcerans_Agy99 MSTARPLRVIQWTTGNIGRRSLHAIIGRPDLELVGVYAHGPDKVGVDAAE
Mb1088|M.bovis_AF2122/97 --MTMSLRVIQWATGSVGVAAIKGVLQHPELELVGCWVHSAAKSGKDVGE
Rv1059|M.tuberculosis_H37Rv --MTMSLRVIQWATGSVGVAAIKGVLQHPELELVGCWVHSAAKSGKDVGE
**.* **.:* : : .* **.* . * * *..*
MMAR_5250|M.marinum_M LCGMA-PLGITASNDVEALLALKPDCVVYNPMWIDVDELVRILSAGVNVV
MAV_0371|M.avium_104 LVGIP-PLGVAATNDVDELLALKPDCVVYNPMWIDVDELVRILSAGVNVV
MSMEG_0560|M.smegmatis_MC2_155 LCGIE-PLGVSATDDVDALLALKPDVVVYNPMWIDVDELVRILESGVNVV
Mflv_1277|M.gilvum_PYR-GCK LVGIE-PLGVAATNDVDALLALKPDAVVYNPMWIDVDELVRILEAGVNVV
Mvan_5538|M.vanbaalenii_PYR-1 LAGIE-PVGVTATNNVDALLALKPDVVVYNPMWIDVDELVRILEAGVNVV
TH_4249|M.thermoresistible__bu LAGIE-PLGVKATNDVDELLALKPDVVVYNPMWIDVDELVRILSAGINVV
MAB_0727|M.abscessus_ATCC_1997 LVGLE-PNGVIATGTVEEIVAAKPDVLTFHGVFPDEDLYVKVLEAGIDIV
MUL_3685|M.ulcerans_Agy99 LAGWPDPTGVRATSDIGALLALGADACCYNPLWPNIDELVGLLESGVNVC
Mb1088|M.bovis_AF2122/97 IIGSP-PLGVIATNSIDDVLALDADAVIYAPLLPSVDEVAALLRSGKNVV
Rv1059|M.tuberculosis_H37Rv IIGSP-PLGVIATNSIDDVLALDADAVIYAPLLPSVDEVAALLRSGKNVV
: * * *: *:. : ::* .* : : . * . :* :* ::
MMAR_5250|M.marinum_M TTAAFITGHNLG------AGR---DRILAACRHGGSTIFGSGVSPG-FAE
MAV_0371|M.avium_104 TTASFITGGNLG------DGR---DRILQACQQGGSTIFGSGVSPG-FAE
MSMEG_0560|M.smegmatis_MC2_155 STASFITGHNQG------TGR---DRIAEACTRGGSSMFGSGISPG-YVN
Mflv_1277|M.gilvum_PYR-GCK ASASFITGHNLG------DGR---AKLADACERGGSTLFGSGVSPG-FAE
Mvan_5538|M.vanbaalenii_PYR-1 ASASFITGRNLG------DGR---DKLADACQRGGSTLFGSGVSPG-FAE
TH_4249|M.thermoresistible__bu SSASFITGHNLR------DGR---DRIEEACRQGGSTMFGSGISPG-FAE
MAB_0727|M.abscessus_ATCC_1997 TTADWITGHHRDQNHPHPSGRPTTEVLQQACERGGSTFYGTGMNPG-LTQ
MUL_3685|M.ulcerans_Agy99 TSAAWITGGKQTP-----RDR---ERILAACERGGATIFGSDAHPG-MTN
Mb1088|M.bovis_AF2122/97 TPLGWFYPSEKE-----------AAPLEVAAQAGNATLHGAGIGPGAVTE
Rv1059|M.tuberculosis_H37Rv TPLGWFYPSEKE-----------AAPLEVAAQAGNATLHGAGIGPGAVTE
:. :: . : *. *.:::.*:. ** .:
MMAR_5250|M.marinum_M LLAIVSAMVCNRIDKVTVNEAADTTFYDSPATER-PVGFGQPIDHPQLQA
MAV_0371|M.avium_104 LLAIVSAMVCNRIDKVTVNEAADTTFYDSPETEK-PVGFGQPIDHPDLPA
MSMEG_0560|M.smegmatis_MC2_155 QLSVVAAGICDRVDKITVNEAADTTFYDSPATEK-PVGFGQPIDHPDLQA
Mflv_1277|M.gilvum_PYR-GCK LLAIVAGTACDRIDKVVISESADTTLYDSPDTER-PVGFDMAIDDPALQP
Mvan_5538|M.vanbaalenii_PYR-1 LLAIVAGTACDRIDKITIAESADTTLYDSPDTER-PVGFGTAIDDPNLQP
TH_4249|M.thermoresistible__bu LLAIIGTQMCDRVDKVTICEQSDTTFYDSPATEK-PVGFGTPIDDPNLPQ
MAB_0727|M.abscessus_ATCC_1997 ILGVIHSCDVAEIENVTVIESVDVSCHHSVDSWE-MVGYGLPVGDPSIPA
MUL_3685|M.ulcerans_Agy99 MVGMVLSGSCERVDEIRITESVDCSTYESAATQT-AMGFSQHPDTPGLAE
Mb1088|M.bovis_AF2122/97 LFPLLLSVMSTGVTFVRSEEFSDLRSYGAPDVLRYVMGFGGTPDSALTGP
Rv1059|M.tuberculosis_H37Rv LFPLLLSVMSTGVTFVRSEEFSDLRSYGAPDVLRYVMGFGGTPDSALTGP
. :: : : * * : : :*:. . .
MMAR_5250|M.marinum_M MTAQGTAIFGEAVRLVADALGVELD-EVRCVAEHAQTTADLDLGSWTIAA
MAV_0371|M.avium_104 MAAKGTAIFGEAVRLVGDALGVELD-DVRCVAEFAQTTEDLVMASWTIPA
MSMEG_0560|M.smegmatis_MC2_155 MTAQGTGVFGEAVRMIADALGIELD-EVRCDAEYAQTTEDLDLGSWTIPA
Mflv_1277|M.gilvum_PYR-GCK MAAQGTAVFAEAVQLVADALGIELD-EITCVSEYAQTTEDLPMASWTIKA
Mvan_5538|M.vanbaalenii_PYR-1 MAANGTAVFAEAVQLVADALAIELD-EIKCVSEYAQTTEDLVMASWTIPA
TH_4249|M.thermoresistible__bu MASQGTAIFAEAVQLIADSLGVELD-EIVCEAEYAQTTEDLDLGSWKIGA
MAB_0727|M.abscessus_ATCC_1997 LLEKGTRVFADGVYLMADCFGLELD-DVTFSYELGACTEDIDLGWYRLPK
MUL_3685|M.ulcerans_Agy99 SVRVESEVFAESAAMMADAIGATLD-KMTFDVTFTAATADTDLGFMKIPA
Mb1088|M.bovis_AF2122/97 MQKILDGGFLQSVRLCVDRLGFAADPQIRTSQEVAVATAPIDSPIGVIEP
Rv1059|M.tuberculosis_H37Rv MQKILDGGFLQSVRLCVDRLGFAADPQIRTSQEVAVATAPIDSPIGVIEP
* :.. : * :. * .: * :
MMAR_5250|M.marinum_M GCVAGVHASWQGVAGGRTIVELNVRWRKG-QTLEPDW--QIDQDGWVIQV
MAV_0371|M.avium_104 GHVAGTYISWQGIVGDQVLIDLNVRWRKG-QTLDPDW--KIEQDGWVIQI
MSMEG_0560|M.smegmatis_MC2_155 GCVAGVYASWKGIVAGHTRVEINVRWRKG-QTLQPDW--KIDQDGWVIEV
Mflv_1277|M.gilvum_PYR-GCK GHVAGVFASWQGIYQGRTVIDINVRWKKG-QTLEPDW--QLDGDGWKITI
Mvan_5538|M.vanbaalenii_PYR-1 GHVAGVYASWQGIANGKTVIDINVRWKKG-QTLDPDW--QLDGDGWKITI
TH_4249|M.thermoresistible__bu GCVAGVYASWQGRIGGRTLIDLNVRWKKG-QTLEPNW--QLEREGWKITV
MAB_0727|M.abscessus_ATCC_1997 GSLGANYLKYQGIVGGVPKVEVHVEWQMT-PKTEPHW--NVKG-CYITKI
MUL_3685|M.ulcerans_Agy99 GTVGSVYGYHRGWVGDRNVVSVGFNWTMG-NHVVPPK--PLKH-GHVIQV
Mb1088|M.bovis_AF2122/97 GQVAGRRFHWEALVEDTVVVQIAVNWLMGSENLDPPWSFGPAGERYEIEV
Rv1059|M.tuberculosis_H37Rv GQVAGRRFHWEALVEDTVVVQIAVNWLMGSENLDPPWSFGPAGERYEIEV
* :.. .. . :.: ..* * :
MMAR_5250|M.marinum_M DGTPTVTTKVSFLPPPYFQ--AETIADFMVLGHIMTAMPTINAIPAVVAA
MAV_0371|M.avium_104 DGQPTVTTKVGFLPPPYFE--ATTIEEFMDLGHIMTAMPAINAIPAVVAA
MSMEG_0560|M.smegmatis_MC2_155 AGRPTVTMKVGFLPPPDFE--ATTLEEFMVLGHIMTATPPLNAIPAVVAA
Mflv_1277|M.gilvum_PYR-GCK EGRPTVNMSVGFLPPQDMIENAKTLEDFFVLGHIMTALPPIHAIPAVVAA
Mvan_5538|M.vanbaalenii_PYR-1 DGRPTVNMQVGFLPPQDMIENAKTLEDFFVLGHIMTAMPPIHAIPAVVAA
TH_4249|M.thermoresistible__bu DGRPTVNMEIGFLPPPDFG--ATTMEEFMVLGHIMTAMPPIHMIPAVVAA
MAB_0727|M.abscessus_ATCC_1997 QGDPCVYSKHMVIPKPGTD--FTDPAAFASVGMTVTGLPALNAIRSVIDA
MUL_3685|M.ulcerans_Agy99 FGLPNMRTVLHCLPPKDWT-----ESGFMGLGMIYTAMPVTNAVPAVVAA
Mb1088|M.bovis_AF2122/97 RGSPDTCVTIKGWQPQTVA-----AGLKSNPGIVATAAHCVNAIPATCAA
Rv1059|M.tuberculosis_H37Rv RGSPDTCVTIKGWQPQTVA-----AGLKSNPGIVATAAHCVNAIPATCAA
* * * *. : : :. *
MMAR_5250|M.marinum_M APGIVTYSDLALTLPRGMVPTDRNSDPKLGASRTP
MAV_0371|M.avium_104 APGIASYADLPLTLPRGNAHVAGQP----------
MSMEG_0560|M.smegmatis_MC2_155 PPGIVTYNDLPLILPRGVVPTP-------------
Mflv_1277|M.gilvum_PYR-GCK APGIATYNDLPLPLPRGVVPTR-------------
Mvan_5538|M.vanbaalenii_PYR-1 APGIATYNDLPLPTPRGVVPTA-------------
TH_4249|M.thermoresistible__bu EPGIATYNDLPLPQARGVVRV--------------
MAB_0727|M.abscessus_ATCC_1997 PPGILTSADLPLRGFAGRFK---------------
MUL_3685|M.ulcerans_Agy99 KPGIVTLADLPPITGRAAL----------------
Mb1088|M.bovis_AF2122/97 PAGIQSFFDLPLITGRAAPGLAR------------
Rv1059|M.tuberculosis_H37Rv PAGIQSFFDLPLITGRAAPGLAR------------
.** : **. .