For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TIDAWMQHPTQRFLRSDMLASLRRWTDSSIPEAQIPIEATIASMDAADVEFGLLSAWRGPNGQDLVSNDE VAGWVDLHPQRFAGLAAVDLDRPMVAVRELRSRIASGFVGLRVVPWLWEAPPTDRRYYPLFAECVESGVP FCTQVGHTGPLRPSETGRPIPYIDQVALDFPELVIVCGHVGYPWTEEMVAVARKHENVYIDTSAYTIKRL PHELIRFMKTGTGRRKVMFGTNFPMIAHAHALAGLDELGLEDEARRDFLHGNAERVFGLAQRPPR*
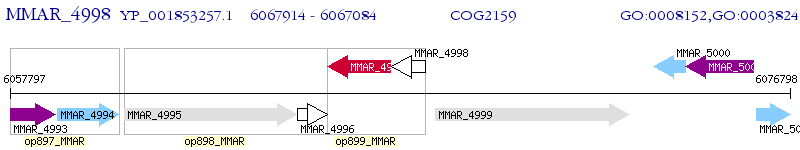
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4998 | - | - | 100% (276) | hypothetical protein MMAR_4998 |
| M. marinum M | MMAR_4012 | - | 8e-25 | 32.16% (199) | metal-dependent hydrolase |
| M. marinum M | MMAR_0140 | - | 2e-21 | 29.75% (242) | hypothetical protein MMAR_0140 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3540c | - | 1e-138 | 84.62% (273) | hypothetical protein Mb3540c |
| M. gilvum PYR-GCK | Mflv_4348 | - | 5e-27 | 35.57% (194) | amidohydrolase 2 |
| M. tuberculosis H37Rv | Rv3510c | - | 1e-138 | 84.62% (273) | hypothetical protein Rv3510c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4667 | - | 7e-12 | 27.68% (177) | putative amidohydrolase |
| M. avium 104 | MAV_0646 | - | 1e-139 | 87.08% (271) | amidohydrolase family protein |
| M. smegmatis MC2 155 | MSMEG_2232 | - | 2e-28 | 32.11% (246) | amidohydrolase family protein |
| M. thermoresistible (build 8) | TH_1218 | - | 2e-27 | 35.18% (199) | amidohydrolase family protein |
| M. ulcerans Agy99 | MUL_3876 | - | 1e-24 | 32.16% (199) | metal-dependent hydrolase |
| M. vanbaalenii PYR-1 | Mvan_1996 | - | 1e-26 | 34.63% (205) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3540c|M.bovis_AF2122/97 -----------MTIDVWMQHPTQRFLHGDMFASLRR---WTGGSIPETDI
Rv3510c|M.tuberculosis_H37Rv -----------MTIDVWMQHPTQRFLHGDMFASLRR---WTGGSIPETDI
MAV_0646|M.avium_104 -----------MTIDVWMQHPTVRFLRSDMLASLRR---WTGGSIPDTDI
MMAR_4998|M.marinum_M -----------MTIDAWMQHPTQRFLRSDMLASLRR---WTDSSIPEAQI
TH_1218|M.thermoresistible__bu ----MTR----KVLDCLVNVHFGEAEMQPAWMEKVRDDYFKGPEAMFAPV
MUL_3876|M.ulcerans_Agy99 ----MTD----RAIDCLVNVHFGEAEAQPNWMLKVRDDYFKGPDSMFAPV
Mflv_4348|M.gilvum_PYR-GCK ----MSD----RVIDCLVNVHFGETANQPEFMLKVRDDYFKGPQSLYDQV
Mvan_1996|M.vanbaalenii_PYR-1 ----MTH----KVIDCLVNVHFGETAKQPEFMLKVRDDYFKGPQSLYEQV
MSMEG_2232|M.smegmatis_MC2_155 ----MTESPAGRAIDCLANVHFGETQNQPAFMTKVRDDYFKGPKSMYDPI
MAB_4667|M.abscessus_ATCC_1997 MSESAFDPAPVRQIWQELGIPGIVDVHTHFMPKSVMDKVWAYFDSAGPLV
: : . :
Mb3540c|M.bovis_AF2122/97 PIEATVSSMDAGGVTLGLLSAW--RGPNGQDLISNDAVAEWVRLYPNRFA
Rv3510c|M.tuberculosis_H37Rv PIEATVSSMDAGGVTLGLLSAW--RGPNGQDLISNDAVAEWVRLYPNRFA
MAV_0646|M.avium_104 PIEATLASMDAAGVGYGLLSAW--RGPNGQDLVSNDEVAQWVAAHPNRFA
MMAR_4998|M.marinum_M PIEATIASMDAADVEFGLLSAW--RGPNGQDLVSNDEVAGWVDLHPQRFA
TH_1218|M.thermoresistible__bu DLAELLDEMDEQGVQKAILMDN--LSK------PSVTARKFVEARPDRFA
MUL_3876|M.ulcerans_Agy99 DLAQLLDEMDEQGVQRAILMDS--LAS------PSLTACKFVQAKPDRFA
Mflv_4348|M.gilvum_PYR-GCK ELPALLDEMAEHGVEKAILMDN--LVK------PSVTARKFAEERPDKFA
Mvan_1996|M.vanbaalenii_PYR-1 ELPALLDEMAEHGVEKAILMDN--LVK------PSVTARKFVEERPDKFA
MSMEG_2232|M.smegmatis_MC2_155 DLGALLEEMDANGVERAILMDN--LTK------PSVTARKFVEAHPERFA
MAB_4667|M.abscessus_ATCC_1997 GRPWPITYRADESQRVTTLRDFGVLRFTSLVYPHKPQMAAWLNQWAAQFA
: . * . : . :**
Mb3540c|M.bovis_AF2122/97 G-LAAVDLDRPMAAVRELRRRVGEG-FVGLRVVPWLWG---APPTDRRYY
Rv3510c|M.tuberculosis_H37Rv G-LAAVDLDRPMAAVRELRRRVGEG-FVGLRVVPWLWG---APPTDRRYY
MAV_0646|M.avium_104 G-LAAVDLDRPMEAVRELRCRIADGGFVGLRVVPWLWN---APPTDRRYY
MMAR_4998|M.marinum_M G-LAAVDLDRPMVAVRELRSRIASG-FVGLRVVPWLWE---APPTDRRYY
TH_1218|M.thermoresistible__bu LAMGGVNLLRPIPSLRELAAVAADLPVVYAVVGPSFWGDGQYPPSDAVYY
MUL_3876|M.ulcerans_Agy99 LAMGGINLLRPVRPLRELTAIVGELPVAYAAVGPSFWGEGQYPPSDAVYY
Mflv_4348|M.gilvum_PYR-GCK LAIGGVNLLRPMPSLRELSSVVADLPVAYAAVGPSFWGDGMYPPSDAVYY
Mvan_1996|M.vanbaalenii_PYR-1 LAVGGVNLLRPMPSLRELAAVVADLPVAYAAVGPSFWGDGMYPPSDAVYY
MSMEG_2232|M.smegmatis_MC2_155 LAMGGVNLLRPIPSLRELAAAVRDLPVAYAVVGPSFWGDGQYPPSDAVYY
MAB_4667|M.abscessus_ATCC_1997 AQTPDCIHTATFFPEPGAADYVREAIDAGVGVFKVHVQVGAFSPTDPLLV
.. . . . * .*:*
Mb3540c|M.bovis_AF2122/97 PLFAECVQSAVPFCTQVGHTGPLRPSETGRPIPYIDQVALDFPELVIVCG
Rv3510c|M.tuberculosis_H37Rv PLFAECVQSAVPFCTQVGHTGPLRPSETGRPIPYIDQVALDFPELVIVCG
MAV_0646|M.avium_104 PLFAACVEAGVPFCTQVGHTGPLRPSETGRPIPYIDQVALDFPELVIVCG
MMAR_4998|M.marinum_M PLFAECVESGVPFCTQVGHTGPLRPSETGRPIPYIDQVALDFPELVIVCG
TH_1218|M.thermoresistible__bu PLYTKCAELELPLCVNTGIPGPPIPGEVQNPI-HLDRVCVRFPELRLCMI
MUL_3876|M.ulcerans_Agy99 PLYVKCAELELALCINTGIPGPPIPGEVQHPM-HLDRVCVRFPELRLCMI
Mflv_4348|M.gilvum_PYR-GCK PLYTKCAELGLPLCINTGLPGPPIPGEVQNPI-HLDRVCVRFPELKLCMI
Mvan_1996|M.vanbaalenii_PYR-1 PLYTKCAELELPLCINTGLPGPPIPGEVQNPI-HLDRVCVRFPELKLCMI
MSMEG_2232|M.smegmatis_MC2_155 PLYTKCAELDLPLCVNTGLPGPPIPGEVQNPI-HLDRVCVRFPELRLCMI
MAB_4667|M.abscessus_ATCC_1997 PVWGLIEDAGVPVVIHCGS-GP-APGEFTGPD-PVTALLKQFPHLRLIVA
*:: : :.. : * ** *.* * : : **.* :
Mb3540c|M.bovis_AF2122/97 HVGYPWTEEMVAVARKHENVYIDT----SAYTIKRLPGKLVRFMKTDTGQ
Rv3510c|M.tuberculosis_H37Rv HVGYPWTEEMVAVARKHENVYIDT----SAYTIKRLPGKLVRFMKTDTGQ
MAV_0646|M.avium_104 HVGYPWTEEMVAVARKHENVYIDT----SAYTIERLPDELVRFIKTGTGQ
MMAR_4998|M.marinum_M HVGYPWTEEMVAVARKHENVYIDT----SAYTIKRLPHELIRFMKTGTGR
TH_1218|M.thermoresistible__bu HGADPWWDIAIRLLIKYQNLRLMT----SAWSPKRLPNSLLHYMRT-RGP
MUL_3876|M.ulcerans_Agy99 HGADPWWDVAIRLLLKYSNSRLMT----SAWTPKRLPESLLHYMRT-RGK
Mflv_4348|M.gilvum_PYR-GCK HGADPWWDVAIRLLIKYANLRIMT----SAWSPKRLPDSLLHYMRT-RGK
Mvan_1996|M.vanbaalenii_PYR-1 HGADPWWDVAIRLLIKYANLRIMT----SAWSPKRLPDSLLHFMRT-RGK
MSMEG_2232|M.smegmatis_MC2_155 HGADPWWDIAIRMLIKYKNLRLMT----SAWSPKRLPESLLHYMRT-RGP
MAB_4667|M.abscessus_ATCC_1997 HMGMPEYSEFLDLCEQHDEVRLDTTMAFTAFSNEKAPFPESDYPRLVEMG
* . * . : : :: : : * :*:: :: * : :
Mb3540c|M.bovis_AF2122/97 RKVLFGTNYPMIAH--THALTGLDELG-LSDEARRDFLHGNAVRVFKLDP
Rv3510c|M.tuberculosis_H37Rv RKVLFGTNYPMIAH--THALTGLDELG-LSDEARRDFLHGNAVRVFKLDP
MAV_0646|M.avium_104 RKVLFGTNYPMITA--EHALAGLDGLG-LSDEARRDFLHGNAERVFRLEA
MMAR_4998|M.marinum_M RKVMFGTNFPMIAH--AHALAGLDELG-LEDEARRDFLHGNAERVFGLAQ
TH_1218|M.thermoresistible__bu NKVIFGSDWPVLTM--RRVVPEAYALD-LPSEVLDNYLYNNAAEFFFGGP
MUL_3876|M.ulcerans_Agy99 NKVIFASDWPVLRM--RRVVPEARALD-LPADVLANYLYNNAQEFFFG-E
Mflv_4348|M.gilvum_PYR-GCK NKVIFASDWPVLRQ--SRVVPEALALD-LPPDVLDNYLYNNANDFFFGTD
Mvan_1996|M.vanbaalenii_PYR-1 NKVIFASDWPVLRQ--SRVVPEAVALD-LPPDVLDNYLYNNANDFFFGAA
MSMEG_2232|M.smegmatis_MC2_155 DKIIFASDFPVLRM--NRVVPEARALD-LPPDVLDNYLYHNAAEFFFGGR
MAB_4667|M.abscessus_ATCC_1997 HKIYFGSDFPNIPYGYAEAITALRRMPGIDDAWMRDVLYHNGARLFGVDT
*: *.:::* : ..:. : : : *: *. .*
Mb3540c|M.bovis_AF2122/97 RGKVQT
Rv3510c|M.tuberculosis_H37Rv RGKVQT
MAV_0646|M.avium_104 AT----
MMAR_4998|M.marinum_M RPPR--
TH_1218|M.thermoresistible__bu EQER--
MUL_3876|M.ulcerans_Agy99 ESGR--
Mflv_4348|M.gilvum_PYR-GCK EQEI--
Mvan_1996|M.vanbaalenii_PYR-1 EQEI--
MSMEG_2232|M.smegmatis_MC2_155 QPQEF-
MAB_4667|M.abscessus_ATCC_1997 SASGR-