For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGWSLASPRLPARWRVALQAVLGGLLVLVTRAPLGLTPPRLWAGLRLGAVAAAPVATAIAATTPVPMVRL SMSERDLPASVPGWLGLQIPFGTVWAEEAAFRAALATMGTRGFGHVGGRLLQAATFGLSHVADARATGAP PVPTVLVTGLAGWLFGWLAERSGSLAAPMVLHLAINEAGAVAALTVQRRWINGAEITPRRRYT*
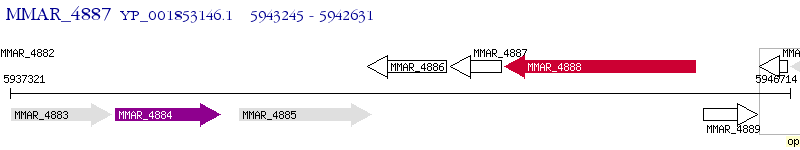
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4887 | - | - | 100% (204) | hypothetical protein MMAR_4887 |
| M. marinum M | MMAR_2740 | - | 8e-15 | 35.98% (189) | hypothetical protein MMAR_2740 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0827 | - | 1e-79 | 76.84% (190) | hypothetical protein Mb0827 |
| M. gilvum PYR-GCK | Mflv_1632 | - | 2e-51 | 54.21% (190) | abortive infection protein |
| M. tuberculosis H37Rv | Rv0804 | - | 1e-79 | 76.84% (190) | hypothetical protein Rv0804 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0709 | - | 2e-46 | 50.26% (189) | hypothetical protein MAB_0709 |
| M. avium 104 | MAV_0745 | - | 3e-64 | 61.50% (200) | caax amino protease family protein |
| M. smegmatis MC2 155 | MSMEG_5821 | - | 7e-56 | 55.10% (196) | caax amino protease family protein |
| M. thermoresistible (build 8) | TH_1367 | - | 2e-14 | 34.20% (193) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_0411 | - | 1e-113 | 98.04% (204) | hypothetical protein MUL_0411 |
| M. vanbaalenii PYR-1 | Mvan_5117 | - | 6e-60 | 59.47% (190) | abortive infection protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4887|M.marinum_M -----------------------MVGWSLAS-PRLPARWRVALQAVLGGL
MUL_0411|M.ulcerans_Agy99 -----------------------MVSWSLAS-PRLPARWRVALQAVLGGL
Mb0827|M.bovis_AF2122/97 -----------MSRLRALSLAAGLVGWSLVS-PRLPAPWRIPLQAGLGSV
Rv0804|M.tuberculosis_H37Rv -----------MSRLRALSLAAGLVGWSLVS-PRLPAPWRIPLQAGLGSV
MAV_0745|M.avium_104 ------MRPSRFRRTAALSLAGALVGWSFVG-PRLPAGARMVLQAGMGAA
MSMEG_5821|M.smegmatis_MC2_155 ------MAPMTAAKFRALALAAALTAWNVVVDPRLPERCRPVVRALVGTA
Mflv_1632|M.gilvum_PYR-GCK ----------MRDTVRAVGLAAALLGWSATA-PRLPQEWNPLPHMVFSGA
Mvan_5117|M.vanbaalenii_PYR-1 ----------MRDGVRATALAMLLLGWSLAA-PRLPQRWNPLPQTMFGAG
MAB_0709|M.abscessus_ATCC_1997 -----------MSGARVLVLAVVLVAWPAVL-ERFPQRWRPLIGAGTSTA
TH_1367|M.thermoresistible__bu VVAQLAALSRFRTYVDVAVVVVVLAICNLIAHFTTPWASVATVPAVAVGL
: *
MMAR_4887|M.marinum_M LVLVTRAPLG-----LTPPRLWAGLRLGAVAAAPVATAIAATTPVPMVRL
MUL_0411|M.ulcerans_Agy99 LVLVTRAPLG-----LTPPRLWAGLRLGAVAAAPVATATAATTPVPMVRL
Mb0827|M.bovis_AF2122/97 LVLVTRATMG-----LWPPRLWAGLRLGWAAGAAAATAIAATTPVPMVRL
Rv0804|M.tuberculosis_H37Rv LVLVTRATMG-----LWPPRLWAGLRLGWAAGAAAATAIAATTPVPMVRL
MAV_0745|M.avium_104 LVALTRAPLG-----LHPPRLWAGLRCGAAVAGCAAGAVAATTLVPAVRE
MSMEG_5821|M.smegmatis_MC2_155 LLGLSRAAPGSRRSGLRPPALWSGMRTGSVAAAVVASSVVATTALPAVRT
Mflv_1632|M.gilvum_PYR-GCK VTLLSRPSLG-----LRPAQLRRGVRWGSAAAAPVGLGVAAGLMAPAVRA
Mvan_5117|M.vanbaalenii_PYR-1 VAALAGAPLG-----LRPPMLWRGLQWGSAAVAPVALSVAAATTSPPVRA
MAB_0709|M.abscessus_ATCC_1997 LAGAAGIPLG-----LRPPQLGAGLRLGGIVASAVAAAVGASPALQPVRA
TH_1367|M.thermoresistible__bu VLLVRSRGLGWAELGLSREHWRSGAGYAAAAVALVLSVIAIGALLPWTRP
: * * * . . . . .*
MMAR_4887|M.marinum_M SMSERDLPASVPGWLG--LQIPFGTVWAEEAAFRAALATMGTRGFGHVGG
MUL_0411|M.ulcerans_Agy99 SMSERDLPASVPGWLG--LQIPFGTVWAEEAAFRAALATMGTRGFGHVGG
Mb0827|M.bovis_AF2122/97 SMSARELPASVPVWLV--WHIPGGTVWAEEAAFRGALATIGARAFGRSGG
Rv0804|M.tuberculosis_H37Rv SMSARELPASVPVWLV--WHIPGGTVWAEEAAFRGALATIGARAFGRSGG
MAV_0745|M.avium_104 SMAGREPPAPAPDWLL--LRIPVGTVWSEEAAFRAALTAAGSAAFGRRSA
MSMEG_5821|M.smegmatis_MC2_155 AMRERELPPAVAFWLM--LGIPLGTVWSEEAAFRGALGAVGTGAFGHTGG
Mflv_1632|M.gilvum_PYR-GCK GMAARDLPAAPARWLL--LRIPFATVGPEELAYRAVLGDAATRAFGPRVG
Mvan_5117|M.vanbaalenii_PYR-1 GMVARDLPDPAASWLL--LRIPFGTVWSEEATFRAALGATAERAFGRTAG
MAB_0709|M.abscessus_ATCC_1997 SMRARDIDLHPAVWLG--LHIPVGTVWSEELAFRGVLQPLATEAFGRTAG
TH_1367|M.thermoresistible__bu MFLNNNYATISGALIASMVIIPLQTVIPEELAFRGVLHGALSRAWGFRGV
: .: : ** ** .** ::*..* .:*
MMAR_4887|M.marinum_M RLLQAATFGLSHVADARATGAP------------------PVPTVLVTGL
MUL_0411|M.ulcerans_Agy99 QLLQAVTFGLSHVADARATGAP------------------PVPTVLVTGL
Mb0827|M.bovis_AF2122/97 RILQAGAFGLSHIADARATGEP------------------LVLTVLATGI
Rv0804|M.tuberculosis_H37Rv RILQAGAFGLSHIADARATGEP------------------LVLTVLATGI
MAV_0745|M.avium_104 MLLQAIAFGLSHIADARAIGEP------------------VAGTVAVTAV
MSMEG_5821|M.smegmatis_MC2_155 RLLQATAFGLSHIPDARAAGEP------------------VVGTVLATGA
Mflv_1632|M.gilvum_PYR-GCK KLFTAVVFGLSHVPDARAAGDN------------------VPGTVVVTGA
Mvan_5117|M.vanbaalenii_PYR-1 RLFAATVFGLSHVPDARAAREP------------------ILGTVLVTGT
MAB_0709|M.abscessus_ATCC_1997 SAVQAMVFGLAHIRPARAAGDS------------------IPGTVLVTGM
TH_1367|M.thermoresistible__bu AAAGSLLFGLWHVATSLGLTSSNVGFTRLFGGGVLGTVAGVVLAVIATAV
: *** *: : . :* .*.
MMAR_4887|M.marinum_M AGWLFGWLAERSGSLAAPMVLHLAINEAGAVAALTVQRRWINGAEITPRR
MUL_0411|M.ulcerans_Agy99 AGWLFGWLAERSGSLAAPMVLHLAINEAGAVAALTVQRRWINGAEITPRR
Mb0827|M.bovis_AF2122/97 AGWMFGWLADRSGSLAAPLLTHLAINEAGAVAAVLVQRR----SGISTRL
Rv0804|M.tuberculosis_H37Rv AGWMFGWLADRSGSLAAPLLTHLAINEAGAVAAVLVQRR----SGISTRL
MAV_0745|M.avium_104 GGWFFGWLTDRCGSLAAPMLAHLAVNEAGAIAVLAVRRR--SGRNAAPRI
MSMEG_5821|M.smegmatis_MC2_155 AGWFLDWLMHRTKSLAAPLLVHLALNEAGALAAVWYARK-------SPRP
Mflv_1632|M.gilvum_PYR-GCK AGWVFSWLHESSGSLAAPVLAHLAVNEAGAVAALAVQQRRSGRGRPTGAA
Mvan_5117|M.vanbaalenii_PYR-1 AGWAFSWLYAKSGSLVAPMLAHLAVNEAGAVAALAVQRRRSARGRPADGA
MAB_0709|M.abscessus_ATCC_1997 FGWLLGRLRERSGSVVAPMLAHLALNEAGAVATLYLGRSNVDLSPESASN
TH_1367|M.thermoresistible__bu AGFVFDWLRRRSGSLLAPIALHWSLNGMGALAAALVWHLST---------
*: :. * *: **: * ::* **:*. :
MMAR_4887|M.marinum_M RYT------
MUL_0411|M.ulcerans_Agy99 RYT------
Mb0827|M.bovis_AF2122/97 ---------
Rv0804|M.tuberculosis_H37Rv ---------
MAV_0745|M.avium_104 ---------
MSMEG_5821|M.smegmatis_MC2_155 Q--------
Mflv_1632|M.gilvum_PYR-GCK R--------
Mvan_5117|M.vanbaalenii_PYR-1 RSPGRRSRS
MAB_0709|M.abscessus_ATCC_1997 ---------
TH_1367|M.thermoresistible__bu ---------