For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GTTPKHSVSVAGVVVRDDGRVLVIRREDNGRWEAPGGVLELHESFEDGVRREVLEETGLKVTVQRLTGVY KNLTQGIVALVYRCGPAAGDTHPTAEAREVRWMTKEQVEGEMSPAFAVRVLDAFDEDPHSRAHDGVNVVS G*
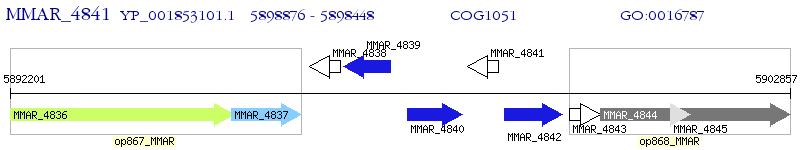
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4841 | - | - | 100% (142) | hypothetical protein MMAR_4841 |
| M. marinum M | MMAR_3129 | - | 1e-11 | 31.71% (123) | hypothetical protein MMAR_3129 |
| M. marinum M | MMAR_4290 | mutT2 | 7e-07 | 34.82% (112) | mutator protein MutT2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3031 | - | 6e-12 | 32.52% (123) | NUDIX hydrolase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2096c | - | 4e-13 | 34.15% (123) | putative MutT/NUDIX-like protein |
| M. avium 104 | MAV_3798 | - | 4e-12 | 32.56% (129) | nudix hydrolase |
| M. smegmatis MC2 155 | MSMEG_5148 | - | 2e-08 | 32.71% (107) | CTP pyrophosphohydrolase |
| M. thermoresistible (build 8) | TH_0645 | mutT2 | 5e-09 | 31.25% (112) | PROBABLE MUTATOR PROTEIN MUTT |
| M. ulcerans Agy99 | MUL_0524 | - | 7e-59 | 75.71% (140) | hypothetical protein MUL_0524 |
| M. vanbaalenii PYR-1 | Mvan_4548 | - | 1e-06 | 33.02% (106) | NUDIX hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4841|M.marinum_M ---------MGTTPKHSVSVAGVVVRDD-GRVLVIRRED----NGRWEAP
MUL_0524|M.ulcerans_Agy99 -------MRMAATPKHSVSVAGIVVRDD-GRVLVIKRDD----NGHWEAP
Mflv_3031|M.gilvum_PYR-GCK MRIDYYNDPNAPAPNSVVPSASAIVTDEHGRILLIKRRD----NTLWALP
MAB_2096c|M.abscessus_ATCC_199 MRTDYYNDPNAPAPNSVVPSASAIVTDEQGRILLIKRRD----NTLWALP
MAV_3798|M.avium_104 MRTDYYNDPNAPRPNSVVPSASAIVADERGRILLIKRRD----NTLWALP
MSMEG_5148|M.smegmatis_MC2_155 ---------MTKQ----IVVAGALISRG--TLLVAQRDRPAELAGLWELP
Mvan_4548|M.vanbaalenii_PYR-1 ---------MSKL----IVVAGALIEGG--ALLVAQRDRPPELAGLWELP
TH_0645|M.thermoresistible__bu ---------MTQIPQPQIVVAGALIADA--ALLVAQRNRPPELAGRWELP
: *. :: :*: :* * *
MMAR_4841|M.marinum_M GGVLELHESFEDGVRREVLEETGLKVTVQRLTGVYKN----------LTQ
MUL_0524|M.ulcerans_Agy99 GGVLELDESFEAGVQREVLEETGVEVTVERLTGVYKN----------LAH
Mflv_3031|M.gilvum_PYR-GCK GGGHDIGETIADTAVREVKEETGLDVEVTGLVGVYTNPHHVVAFTDGEVR
MAB_2096c|M.abscessus_ATCC_199 GGGHDIGETIADTAVREVKEETGLDVEVTGLVGVYTNPHHVVAFTDGEVR
MAV_3798|M.avium_104 GGGHDIGETIEQTAVREVKEETGLDVEITGLVGVYTNPRHVVAFTDGEVR
MSMEG_5148|M.smegmatis_MC2_155 GGKVTPGESDADALARELREELGVDVAVGERLGADVA----------LND
Mvan_4548|M.vanbaalenii_PYR-1 GGKVAPGESDEAALVRELNEELGVDVTVGARVGADIA----------LSA
TH_0645|M.thermoresistible__bu GGKVAPGETEAEALVRELKEELGVDVEVGARLGVEVA----------LTE
** *: **: ** *:.* : *.
MMAR_4841|M.marinum_M GIVALVYRCGPAAGDTHPTAEAREVRWMTKEQVEG-EMSPAFAVRVLDAF
MUL_0524|M.ulcerans_Agy99 GIVALVYRCRPLGDEAHATEEACEIRWMTKEEVQS-AMVPAFGVRVLDAF
Mflv_3031|M.gilvum_PYR-GCK QQFSLSFTTKVLGGTLAIDHESTDIAWTSPDDIPSLDMHPSMRLRIEHYL
MAB_2096c|M.abscessus_ATCC_199 QQFSLLFATTVLGGTLAIDHESTDIAWTAPDDIAGLDMHPSMRLRVEHYL
MAV_3798|M.avium_104 QQFSLLFTTRVLGGELAIDHESTDIAWTDPDDIADLDMHPSMRLRIEHYL
MSMEG_5148|M.smegmatis_MC2_155 AMTLRAYRVTLRSGSPHP-HDHRALRWVGADEIDGLAWVPADRAWVPDLV
Mvan_4548|M.vanbaalenii_PYR-1 AMCLRAYAVTRTRGVVAP-HDHRALRWIRTEEIETLPWVPADRAWLPDLT
TH_0645|M.thermoresistible__bu SMVLRAFRVHHRGGSPSP-NDHRALRWVTAAELHDLDWVPADRAWIPELT
: . : : * :: *: : .
MMAR_4841|M.marinum_M DEDPHSRAHDGVNVVSG-------
MUL_0524|M.ulcerans_Agy99 EEAPQSRAHDGVNLVRV-------
Mflv_3031|M.gilvum_PYR-GCK QHRDNPYLG---------------
MAB_2096c|M.abscessus_ATCC_199 QRRDSPYLG---------------
MAV_3798|M.avium_104 QHRDSPYLC---------------
MSMEG_5148|M.smegmatis_MC2_155 AALSGR------------------
Mvan_4548|M.vanbaalenii_PYR-1 RLLAGADHARASGAAARRGNQPPA
TH_0645|M.thermoresistible__bu ALLSAR------------------