For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRIATLAVSRSTFRKKHLRANDQRQGLGAQIMDLFDKFVAVDEPLLHISRATHGIATYPKLDGPVGAHMR WQDREQIVWSINNYLGIANLPEVREADVEFARLYGFARPMGSRMMCGETDILEQLEEELANYVKKPAALF LNYGYQGMASLIDSLTTSSDWIVLDSQCHACIIDGVRLKKKGGLDQTRTFSHNNIDELQACLAEIDRVRS ADAGVLVITEGVFGMSGDQGALREIVELKKTYDFRLLVDDAHGFGALGATGSGAGEEQGVQDGIDLYFST FAKAAAGTGAFVASEANVIWKLRYTMRSQIFSRGLPLPIIAGNLFRFELLRTRHDLRQRAHAIAHELQTS LTDKGFDIGNTQSLVTPVFLQMDPLLTLEFLNRMRHEHNVFCSGVMYPVVPPGVVQLRLVSTASHDTADV EPTVNAIASLYDELVPNRDRDLSPAPELGAV
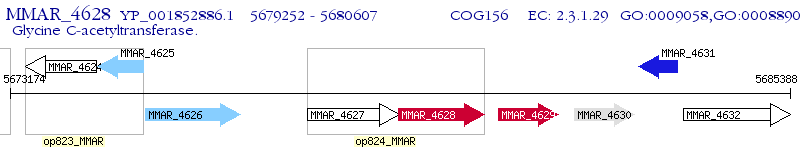
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4628 | bioF2_1 | - | 100% (451) | 8-amino-7-oxononanoate synthase BioF2_1 |
| M. marinum M | MMAR_1724 | bioF2_2 | e-162 | 68.80% (407) | 8-amino-7-oxononanoate synthase BioF2_2 |
| M. marinum M | MMAR_0784 | bioF2_4 | 4e-38 | 29.00% (400) | 8-amino-7-oxononanoate synthase BioF2_4 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0033 | bioF2 | 8e-43 | 31.05% (380) | 8-amino-7-oxononanoate synthase BioF2 |
| M. gilvum PYR-GCK | Mflv_3628 | - | 3e-24 | 27.98% (336) | 8-amino-7-oxononanoate synthase |
| M. tuberculosis H37Rv | Rv0032 | bioF2 | 8e-43 | 31.05% (380) | 8-amino-7-oxononanoate synthase BioF2 |
| M. leprae Br4923 | MLBr_01217 | bioF | 8e-23 | 28.65% (342) | 8-amino-7-oxononanoate synthase |
| M. abscessus ATCC 19977 | MAB_2687c | - | 5e-28 | 31.44% (353) | 8-amino-7-oxononanoate synthase |
| M. avium 104 | MAV_3205 | bioF | 5e-26 | 30.64% (346) | 8-amino-7-oxononanoate synthase |
| M. smegmatis MC2 155 | MSMEG_3189 | bioF | 6e-26 | 29.25% (335) | 8-amino-7-oxononanoate synthase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0241 | bioF2_1 | 0.0 | 98.81% (420) | 8-amino-7-oxononanoate synthase BioF2_1 |
| M. vanbaalenii PYR-1 | Mvan_2789 | - | 6e-23 | 29.60% (277) | 8-amino-7-oxononanoate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3628|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2789|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3189|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01217|M.leprae_Br4923 --------------------------------------------------
MAV_3205|M.avium_104 --------------------------------------------------
MAB_2687c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0033|M.bovis_AF2122/97 MPTGLGYDFLRPVEDSGINDLKHYYFMADLADGQPLGRANLYSVCFDLAT
Rv0032|M.tuberculosis_H37Rv MPTGLGYDFLRPVEDSGINDLKHYYFMADLADGQPLGRANLYSVCFDLAT
MMAR_4628|M.marinum_M --------------------------------------------------
MUL_0241|M.ulcerans_Agy99 --------------------------------------------------
Mflv_3628|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2789|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3189|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01217|M.leprae_Br4923 --------------------------------------------------
MAV_3205|M.avium_104 --------------------------------------------------
MAB_2687c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0033|M.bovis_AF2122/97 TDRKLTPAWRTTIKRWFPGFMTFRFLECGLLTMVSNPLALRSDTDLERVL
Rv0032|M.tuberculosis_H37Rv TDRKLTPAWRTTIKRWFPGFMTFRFLECGLLTMVSNPLALRSDTDLERVL
MMAR_4628|M.marinum_M --------------------------------------------------
MUL_0241|M.ulcerans_Agy99 --------------------------------------------------
Mflv_3628|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2789|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3189|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01217|M.leprae_Br4923 --------------------------------------------------
MAV_3205|M.avium_104 --------------------------------------------------
MAB_2687c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0033|M.bovis_AF2122/97 PVLAGQMDQLAHDDGSDFLMIRDVDPEHYQRYLDILRPLGFRPALGFSRV
Rv0032|M.tuberculosis_H37Rv PVLAGQMDQLAHDDGSDFLMIRDVDPEHYQRYLDILRPLGFRPALGFSRV
MMAR_4628|M.marinum_M --------------------------------------------------
MUL_0241|M.ulcerans_Agy99 --------------------------------------------------
Mflv_3628|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2789|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3189|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01217|M.leprae_Br4923 --------------------------------------------------
MAV_3205|M.avium_104 --------------------------------------------------
MAB_2687c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0033|M.bovis_AF2122/97 DTTISWSSVEEALGCLSHKRRLPLKTSLEFRERFGIEVEELDEYAEHAPV
Rv0032|M.tuberculosis_H37Rv DTTISWSSVEEALGCLSHKRRLPLKTSLEFRERFGIEVEELDEYAEHAPV
MMAR_4628|M.marinum_M --------------------------------------------------
MUL_0241|M.ulcerans_Agy99 --------------------------------------------------
Mflv_3628|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2789|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3189|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01217|M.leprae_Br4923 --------------------------------------------------
MAV_3205|M.avium_104 --------------------------------------------------
MAB_2687c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0033|M.bovis_AF2122/97 LARLWRNVKTEAKDYQREDLNPEFFAACSRHLHGRSRLWLFRYQGTPIAF
Rv0032|M.tuberculosis_H37Rv LARLWRNVKTEAKDYQREDLNPEFFAACSRHLHGRSRLWLFRYQGTPIAF
MMAR_4628|M.marinum_M --------------------------------------------------
MUL_0241|M.ulcerans_Agy99 --------------------------------------------------
Mflv_3628|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2789|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3189|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01217|M.leprae_Br4923 --------------------------------------------------
MAV_3205|M.avium_104 --------------------------------------------------
MAB_2687c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0033|M.bovis_AF2122/97 FLNVWGADENYILLEWGIDRDFEHYRKANLYRAALMLSLKDAISRDKRRM
Rv0032|M.tuberculosis_H37Rv FLNVWGADENYILLEWGIDRDFEHYRKANLYRAALMLSLKDAISRDKRRM
MMAR_4628|M.marinum_M ------------------------------------------MRIATLAV
MUL_0241|M.ulcerans_Agy99 --------------------------------------------------
Mflv_3628|M.gilvum_PYR-GCK ----------------------------MTRAGLSPLAWLDEVETQR---
Mvan_2789|M.vanbaalenii_PYR-1 ----------------------------MTRVSLSPLAWLDEVETQR---
MSMEG_3189|M.smegmatis_MC2_155 ----------------------------MTRAGLSPLAWLADIEQRR---
MLBr_01217|M.leprae_Br4923 ---------------------------MKVPIETSPLAWLEAVEQQR---
MAV_3205|M.avium_104 ---------------------------MKAPIEVSPLAWLDAVEQQR---
MAB_2687c|M.abscessus_ATCC_199 ---------------------------MPVVEASSPLAWLGEVERQR---
Mb0033|M.bovis_AF2122/97 EMGITNYFTKLRIPGARVIPTIYFLRHSTDPVHTATLARMMMHNIQRPTL
Rv0032|M.tuberculosis_H37Rv EMGITNYFTKLRIPGARVIPTIYFLRHSTDPVHTATLARMMMHNIQRPTL
MMAR_4628|M.marinum_M SRSTFRKKHLRANDQRQGLGAQIMDLFDKFVAVDEPLLHISRATHGIATY
MUL_0241|M.ulcerans_Agy99 -----------------------MDLFDKFVAVDEPLLHISRATHGIATY
.* :
Mflv_3628|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2789|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3189|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01217|M.leprae_Br4923 --------------------------------------------------
MAV_3205|M.avium_104 --------------------------------------------------
MAB_2687c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0033|M.bovis_AF2122/97 PDDMSEEFCRWEERIRLDQDGLPEHDIFRKIDRQHKYTGLKLGGVYGFYP
Rv0032|M.tuberculosis_H37Rv PDDMSEEFCRWEERIRLDQDGLPEHDIFRKIDRQHKYTGLKLGGVYGFYP
MMAR_4628|M.marinum_M P-------------------------------------------------
MUL_0241|M.ulcerans_Agy99 P-------------------------------------------------
Mflv_3628|M.gilvum_PYR-GCK RAAGLRRSLRARPPVGTVLDLASNDYLGLSQHPRVIDGGVAALRTWGAG-
Mvan_2789|M.vanbaalenii_PYR-1 RAAGLRRTVRTRPPVGAELDLASNDYLGLSQHPRVIDGGVSALRTWGAG-
MSMEG_3189|M.smegmatis_MC2_155 RAEGLRRELRVRPPVAAELDLASNDYLGLSQHPDVLDGGVEALRTWGGG-
MLBr_01217|M.leprae_Br4923 RGAGLRRSLRPRSAVATELDLASNDYLGLSQHPDVIDGGVAALRVWGAG-
MAV_3205|M.avium_104 RAAGLRRSLRPRPPVATELDLASNDYLGLSQHPDVIDGGVAALRLWGAG-
MAB_2687c|M.abscessus_ATCC_199 EAAGLRRRLRTRSAAEPEIDLASNDYLGLSRHPQVIEAGVEALRIWGAG-
Mb0033|M.bovis_AF2122/97 RFTGPQRSTVKAAELGEIVLLGTNSYLGLATHPEVVEASAEATRRYGTG-
Rv0032|M.tuberculosis_H37Rv RFTGPQRSTVKAAELGEIVLLGTNSYLGLATHPEVVEASAEATRRYGTG-
MMAR_4628|M.marinum_M KLDGPVG-AHMRWQDREQIVWSINNYLGIANLPEVREADVEFARLYGFAR
MUL_0241|M.ulcerans_Agy99 KLDGPVG-AHMRWHDREQIVWSINNYLGIANLPEVREADVEFARLYGFAR
. * : . *.***:: * * :... * :* .
Mflv_3628|M.gilvum_PYR-GCK ATGSRLVTGNTELHEQFEDALASFVGADSALVFSSGYTANLGAVVALSGP
Mvan_2789|M.vanbaalenii_PYR-1 STGSRLVTGNTELHEAFEDVLAAFVGAESALVFSSGYTANLGAVVALSGP
MSMEG_3189|M.smegmatis_MC2_155 AGGSRLVTGNTELHEAFEHQLASFLGAESALVFSSGYTANLGALVALSGP
MLBr_01217|M.leprae_Br4923 ATGSRLVTGDTILHHELECELAEFVGACVGLLFSSGYAANLGAVVGLSGP
MAV_3205|M.avium_104 ATGSRLVTGDTELHQQFESELADYVGAASGLLFSSGYAANLGAVVGLSGR
MAB_2687c|M.abscessus_ATCC_199 STGSRLVTGNTELHEELEQELATFMGTESALVFSSGYTANLGAVVALSGP
Mb0033|M.bovis_AF2122/97 CSGSPLLNGTLDLHVSLEQELACFLGKPAAVLCSTGYQSNLAAISALCES
Rv0032|M.tuberculosis_H37Rv CSGSPLLNGTLDLHVSLEQELACFLGKPAAVLCSTGYQSNLAAISALCES
MMAR_4628|M.marinum_M PMGSRMMCGETDILEQLEEELANYVKKPAALFLNYGYQGMASLIDSLTTS
MUL_0241|M.ulcerans_Agy99 PMGPRMMCGETDILEQLEEELANYVKKPAALFLNYGYQGMASLIDSLTTS
*. :: * : :* ** :: .:. . ** . . : .*
Mflv_3628|M.gilvum_PYR-GCK GSLLVSDAYTHASLVDACRLSR----ARVVVTPHNDVAAVARALATRDEE
Mvan_2789|M.vanbaalenii_PYR-1 GSLLVSDANTHASLVDACRLSR----ARVVVTPHNDTAAVEQALATRSEA
MSMEG_3189|M.smegmatis_MC2_155 GSLIVSDALSHASLVDACRLSR----ARVVVSPHRDVDAVDAALAARTEE
MLBr_01217|M.leprae_Br4923 GSLIVSDAYSHASLVDACRLSR----ARVVVTPHCDVDAVDTALRSCHEE
MAV_3205|M.avium_104 GALVVSDAYSHASLVDACRLSR----ARVVVTPHRDVDAVLAALADRDEE
MAB_2687c|M.abscessus_ATCC_199 GTLIVSDARTHASLIDACRLSR----ARVVVTPYRDTQAVAAALAQRPEE
Mb0033|M.bovis_AF2122/97 GDMIIQDALNHRSLFDAARLSG----ADFTLYRHNDMDHLARVLRRTEGR
Rv0032|M.tuberculosis_H37Rv GDMIIQDALNHRSLFDAARLSG----ADFTLYRHNDMDHLARVLRRTEGR
MMAR_4628|M.marinum_M SDWIVLDSQCHACIIDGVRLKKKGGLDQTRTFSHNNIDELQACLAEIDRV
MUL_0241|M.ulcerans_Agy99 SDWIVLDSQCHACIIDGVRLKKKGGLDQTRTFSHNNIDELQACLAEIDRV
. :: *: * .:.*. **. : : : *
Mflv_3628|M.gilvum_PYR-GCK R-----AVVVTDSVFSADGDLAPLRELHDACRRHGALMIVDEAHGLGVRG
Mvan_2789|M.vanbaalenii_PYR-1 R-----AVVVTDSVFSADGDLAPLRALHTACRKHGALLIVDEAHGLGVRG
MSMEG_3189|M.smegmatis_MC2_155 R-----AVVVTESVFSADGDLAPLRDLHAVCRRHGALLLVDEAHGLGVRG
MLBr_01217|M.leprae_Br4923 R-----AVVVTESVFSADGVLAPVSELHDVCRRHGALLLVDEAHGLGVRG
MAV_3205|M.avium_104 R-----AVVITESVFSTDGALAPLRELHEVCRRHRALLIVDEAHGLGVRG
MAB_2687c|M.abscessus_ATCC_199 R-----ALVLTDAVFSADGALAPLAELYGVCRRHGAVLLVDEAHGLGVRG
Mb0033|M.bovis_AF2122/97 R-----RIIVVDAVFSMEGTVADLATIAELADRHGCRVYVDESHALGVLG
Rv0032|M.tuberculosis_H37Rv R-----RIIVVDAVFSMEGTVADLATIAELADRHGCRVYVDESHALGVLG
MMAR_4628|M.marinum_M RSADAGVLVITEGVFGMSGDQGALREIVELKKTYDFRLLVDDAHGFGALG
MUL_0241|M.ulcerans_Agy99 RSADAGVLVITEGVFGMSGDQGALREIVELKKTYDFRLLVDDAHGFGALG
* :::.:.**. .* . : : : : **::*.:*. *
Mflv_3628|M.gilvum_PYR-GCK DRGRGLLDEVGLAGAPDVVMTTTLSKALGSQGGVVLGPAAVRDHLIDAAR
Mvan_2789|M.vanbaalenii_PYR-1 DGGRGLLDEVGLAGAPDLVMTTTLSKALGSQGGVVLGPAAVRAHLIDTAR
MSMEG_3189|M.smegmatis_MC2_155 TRGQGLLHEVGLAGAPDIVMTTTLSKALGSQGGAVLGPEAVRAHLIDTAR
MLBr_01217|M.leprae_Br4923 -GGRGLVYEVGLAGAPDVVITTTMSKALGSQGGAVLGSSAVRAHLINTAR
MAV_3205|M.avium_104 -GGRGLVFEAGLAGAPDVVMTTTLSKALGSQGGAVLGPAAVRAHLIDAAR
MAB_2687c|M.abscessus_ATCC_199 SGGRGLVAEMNLAGRDDLVVTVTLSKALGSQGGAVLGSAAVRAHLIDAAR
Mb0033|M.bovis_AF2122/97 PDGRGASAALGVLARMDVVMG-TFSKSFASVGGFIAGDRPVVDYIRHNGS
Rv0032|M.tuberculosis_H37Rv PDGRGASAALGVLARMDVVMG-TFSKSFASVGGFIAGDRPVVDYIRHNGS
MMAR_4628|M.marinum_M ATGSGAGEEQGVQDGIDLYFS-TFAKAAAGTGAFVASEANVIWKLRYTMR
MUL_0241|M.ulcerans_Agy99 ATGSGAGEEQGVQDGIDLYFS-TFAKAAADTGAFVASEANVIWKLRYTMR
* * .: *: . *::*: .. *. : . * :
Mflv_3628|M.gilvum_PYR-GCK PFIFDTGLAPAAVGAAHAALQVLVEEPWRAQRVLDHATTLAQICGVA---
Mvan_2789|M.vanbaalenii_PYR-1 PFIFDTGLAPAAVGAAFAALQVLIDEPWRAQRVLDHAATLASICDVT---
MSMEG_3189|M.smegmatis_MC2_155 SFIFDTGLAPAAVGAASAALRVLDAEPQRARAVLDRAAELATIAGVT---
MLBr_01217|M.leprae_Br4923 PFIFDTGLAPAAVGAARAALQILKAETWRPEAVLQHARTLAKICDLS---
MAV_3205|M.avium_104 TFIFDTGLAPAAVGAARAALGVLRAEPWRVGAVLRHAGVLAEVCRVR---
MAB_2687c|M.abscessus_ATCC_199 PFIFDTGLAPAAVGSALAALRLMIDEPARIRAVLAHAAALGQWCGVP---
Mb0033|M.bovis_AF2122/97 GHVFSASLPPAAAAATHAALRVSRREPDRRARVLAAAEYMATGLARQGYQ
Rv0032|M.tuberculosis_H37Rv GHVFSASLPPAAAAATHAALRVSRREPDRRARVLAAAEYMATGLARQGYQ
MMAR_4628|M.marinum_M SQIFSRGLPLPIIAGNLFRFELLRTRHDLRQRAHAIAHELQTSLTDKGFD
MUL_0241|M.ulcerans_Agy99 SQIFSRGLPLPIIAGNLFRFELLRTRHDLRQRAHAIAHELQTSLTDKGFY
:*. .*. . .. : : . . * :
Mflv_3628|M.gilvum_PYR-GCK -DVPSSAVVSVILGEPEVALAAAMGCLERGVRVGC--FRPPTVPAGTSRL
Mvan_2789|M.vanbaalenii_PYR-1 -EVPSSAVVSVILGEPEVAFAAAAACLDRGVRVGC--FRPPTVPAGTSRL
MSMEG_3189|M.smegmatis_MC2_155 -EAPVSAVVSVILGDPEIAVGAAAACLDRGVRVGC--FRPPTVPAGTSRL
MLBr_01217|M.leprae_Br4923 -ELPQSAVVSVVLGDPEVALAAAIACLDAGVRVGC--FRPPTVPAGTSRL
MAV_3205|M.avium_104 -EVPQSAVVSVILGDPDVAVAAATACLDAGVRVGC--FRPPTVPAGTSRL
MAB_2687c|M.abscessus_ATCC_199 -DKPESAVVPVVLGDPTVAFNAARSCLEQGVRVGC--FRPPSVPEGQSLL
Mb0033|M.bovis_AF2122/97 AEYHGTAIVPVILGNPTVAHAGYLRLMRSGVYVNP--VAPPAVPEERSGF
Rv0032|M.tuberculosis_H37Rv AEYHGTAIVPVILGNPTVAHAGYLRLMRSGVYVNP--VAPPAVPEERSGF
MMAR_4628|M.marinum_M IGNTQSLVTPVFLQMDPLLTLEFLNRMRHEHNVFCSGVMYPVVPPGVVQL
MUL_0241|M.ulcerans_Agy99 IGNTQSLVTPVFLQMDPLLTLEFVNRMRHEHNVFCSGVMYPVVPPGVVQL
: :..*.* : : * . * ** :
Mflv_3628|M.gilvum_PYR-GCK RLTARASLSDDEMALAREVLTDVLSRA-----------------
Mvan_2789|M.vanbaalenii_PYR-1 RLTARASLSEDEMTLARRVLTEVLSPR-----------------
MSMEG_3189|M.smegmatis_MC2_155 RLAARASLTDDEMALARQVLTDVLATARA---------------
MLBr_01217|M.leprae_Br4923 RLTAHASLDSAKLEVARRVLTDVLGGCCVARR------------
MAV_3205|M.avium_104 RLTARASLDDAELEVARRVLTDVLAGLG----------------
MAB_2687c|M.abscessus_ATCC_199 RLTARASLTEADMERVHEVLAGVLTLARR---------------
Mb0033|M.bovis_AF2122/97 RTSYLADHRQSDLDRALHVFAGLAEDLTPQGAAL----------
Rv0032|M.tuberculosis_H37Rv RTSYLADHRQSDLDRALHVFAGLAEDLTPQGAAL----------
MMAR_4628|M.marinum_M RLVSTASHDTADVEPTVNAIASLYDELVPNRDRDLSPAPELGAV
MUL_0241|M.ulcerans_Agy99 RLVSTASHDTADVEPTVNAIASLYDELVPNRDRDLSPAPELGAV
* *. .: . ..:: :