For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLFTSDRGIAPAAAARLADDHGFHTFYVPEHTHIPIKREAAHPTTGDSSLPDDRYMRTLDPWVSLGAASA VTSRVRLSTAVALPVEHDPITLAKSIATLDHLSGGRVSLGVGFGWNTDELADHGVPPGRRRTMLREYLEA MRALWTQEEAAYDGEFVKFGPSWAWPKPVQSHIPVLVGAAGTEKNFKWIARSADGWITTPRDFDIDAPVK LLQDTWAAAGRDGAPQIVALDFKPVPEKLARWAELGVTEVLFGLPDRPEDEIAAYVERLAGKLAAMV
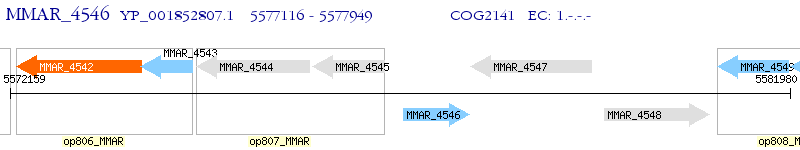
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4546 | - | - | 100% (277) | oxidoreductase |
| M. marinum M | MMAR_3559 | - | 6e-49 | 40.22% (276) | hypothetical protein MMAR_3559 |
| M. marinum M | MMAR_1575 | - | 6e-48 | 39.13% (276) | oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0978c | - | 1e-148 | 89.89% (277) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1871 | - | 1e-149 | 91.34% (277) | luciferase family protein |
| M. tuberculosis H37Rv | Rv0953c | - | 1e-148 | 89.89% (277) | oxidoreductase |
| M. leprae Br4923 | MLBr_00348 | - | 8e-12 | 30.67% (150) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_1059c | - | 1e-125 | 74.18% (275) | hypothetical protein MAB_1059c |
| M. avium 104 | MAV_1077 | - | 1e-154 | 94.58% (277) | hypothetical protein MAV_1077 |
| M. smegmatis MC2 155 | MSMEG_5520 | - | 1e-146 | 88.81% (277) | hypothetical protein MSMEG_5520 |
| M. thermoresistible (build 8) | TH_2830 | - | 1e-146 | 90.11% (273) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_3458 | - | 2e-47 | 38.77% (276) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_4864 | - | 1e-149 | 90.58% (276) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0978c|M.bovis_AF2122/97 -------------------MHYGLVLFTSDRGITPAAAARLAESHGFRTF
Rv0953c|M.tuberculosis_H37Rv -------------------MHYGLVLFTSDRGITPAAAARLAESHGFRTF
MMAR_4546|M.marinum_M ------------------------MLFTSDRGIAPAAAARLADDHGFHTF
MAV_1077|M.avium_104 ------------------------MLFTSDRGISPAAAAKLADDHGFATF
Mflv_1871|M.gilvum_PYR-GCK MQGWHPPCGRGLEHVLILGMNYGLVLFTSDRGITPAAAATLADQHGFETF
Mvan_4864|M.vanbaalenii_PYR-1 -------------------MNYGLVLFTSDRGITPAAAAKLADEHGFETF
MSMEG_5520|M.smegmatis_MC2_155 -------------------MDYGLVLFTSDRGITPASAAKLADDHGFTTF
TH_2830|M.thermoresistible__bu -------------------MDFGLVLFTSDRGITPAKAATLADDHGFTTF
MAB_1059c|M.abscessus_ATCC_199 -------------------MDYGLVLFTSDRGISPAIAAKAAEDGGFSTF
MUL_3458|M.ulcerans_Agy99 -------------------MRVGVLTFVTDEGIGPAELGAALEERGFESL
MLBr_00348|M.leprae_Br4923 -------------------------------MVERANAMKLGLQLGYWAA
: * . *: :
Mb0978c|M.bovis_AF2122/97 YVPEHTHIPVKRQAAHPTTGDASLPDDRYMRTLDPWVSLGAASAVTSRIR
Rv0953c|M.tuberculosis_H37Rv YVPEHTHIPVKRQAAHPTTGDASLPDDRYMRTLDPWVSLGAASAVTSRIR
MMAR_4546|M.marinum_M YVPEHTHIPIKREAAHPTTGDSSLPDDRYMRTLDPWVSLGAASAVTSRVR
MAV_1077|M.avium_104 YVPEHTHIPIKREAAHPTTGDASLPDDRYMRTLDPWVSLGAACAVTSRVR
Mflv_1871|M.gilvum_PYR-GCK YVPEHTHIPIKRQAAHPTTGDESLPDDRYMRTLDPWVSLGTAAAVTSRVR
Mvan_4864|M.vanbaalenii_PYR-1 YVPEHTHIPIKRQAAHPTTGDESLPDDRYMRTLDPWVSLGTAAAVTSRVR
MSMEG_5520|M.smegmatis_MC2_155 YVPEHTHIPIKREAAHPTTGDESLPDDRYMRTLDPWVSLGTAAAVTSRVG
TH_2830|M.thermoresistible__bu YVPEHTHIPVKREAAHPTTGDSSLPDDRYMRTLDPWVSLATASAVTSRVR
MAB_1059c|M.abscessus_ATCC_199 YVPEHTHIPVKREAAHPGTGGAELPDDRYMRTLDPWVSLGAACAVTSKIR
MUL_3458|M.ulcerans_Agy99 FLAEHTHIPVNNQSPYPPGG--PIPR-KYYRTLDPFVALTAAAVATESLV
MLBr_00348|M.leprae_Br4923 QPPGN---AAELVAAAEGAGFDAVFTGEAWG-SDAYTPLAWWGSSTQRVR
. : . : :. * : . *.:..* *. :
Mb0978c|M.bovis_AF2122/97 LATAVALPVEHDPITLAKSIATLDHLSHGRVSVGVGFGWNTDELVDHGVP
Rv0953c|M.tuberculosis_H37Rv LATAVALPVEHDPITLAKSIATLDHLSHGRVSVGVGFGWNTDELVDHGVP
MMAR_4546|M.marinum_M LSTAVALPVEHDPITLAKSIATLDHLSGGRVSLGVGFGWNTDELADHGVP
MAV_1077|M.avium_104 LSTAVALPVEHDPITLAKSIATLDHLSGGRVSLGVGFGWNTDELADHGVP
Mflv_1871|M.gilvum_PYR-GCK LSTAVALPVEHDCITLAKSIATLDHLSGGRVSLGVGFGWNTDELADHNVP
Mvan_4864|M.vanbaalenii_PYR-1 LSTAVALPVEHDPITLAKSIATLDHLSGGRVSLGVGFGWNTDELADHNVP
MSMEG_5520|M.smegmatis_MC2_155 LSTAVALPVEHDPITLAKSIATLDHLSGGRVSLGVGFGWNTDELADHKVP
TH_2830|M.thermoresistible__bu LSTAVALPVEHDPISLAKTIASLDHLSGGRVSLGVGFGWNTDELVDHGVP
MAB_1059c|M.abscessus_ATCC_199 LATAVAIPVEHDPITLAKSIATLDHMSGGRVTVGVGYGWNTDELADHHVP
MUL_3458|M.ulcerans_Agy99 LGTGIALIPERDPIVTAKEVASLDVVSGGRFCFGVGVGWLREEVANHGVD
MLBr_00348|M.leprae_Br4923 LGTSVVQLSARTPTACAMAALTLDHLSGGRHILGLGVSGPQVVEGWYGQP
*.*.:. : * :** :* ** .*:* . :
Mb0978c|M.bovis_AF2122/97 PGRRRTMLREYLEAMRALWTQEE-ACYDGEFVKFGPSWAWPK--------
Rv0953c|M.tuberculosis_H37Rv PGRRRTMLREYLEAMRALWTQEE-ACYDGEFVKFGPSWAWPK--------
MMAR_4546|M.marinum_M PGRRRTMLREYLEAMRALWTQEE-AAYDGEFVKFGPSWAWPK--------
MAV_1077|M.avium_104 AGRRRTMLREYLEAMRALWTQEE-AAYDGEFVKFGPSWAWPK--------
Mflv_1871|M.gilvum_PYR-GCK PGRRRTMLREYLEAMRALWTQEE-AEYDGEFVKFGPSWAWPK--------
Mvan_4864|M.vanbaalenii_PYR-1 PGRRRTMLREYLEAMRALWTQEE-AEFDGEFVKFGPSWAWPK--------
MSMEG_5520|M.smegmatis_MC2_155 AGRRRTMLREYLEAMRALWTQEE-ASYDGEFVNFGPSWAWPK--------
TH_2830|M.thermoresistible__bu PKRRRTMLREYLEAMRALWTQEE-ASYQGEFVSFGPSWAWPK--------
MAB_1059c|M.abscessus_ATCC_199 AGRRRTMLREYIEAMRRLWEEEE-ASYSGEFVNFGPSWAWPK--------
MUL_3458|M.ulcerans_Agy99 PAVRGRVSDERLGAMIEIWTQDN-AEYHGKYVDFDPICCWPK--------
MLBr_00348|M.leprae_Br4923 FPKPLSRTREYIDIVRQVWAREAPVVSDGQHYPLPLTGAGATGLGKALKP
* : : :* .: . *:. : . ..
Mb0978c|M.bovis_AF2122/97 ---PVQPHIPVLVGAAGTEKNFKWIARSADGWITTPR-------------
Rv0953c|M.tuberculosis_H37Rv ---PVQPHIPVLVGAAGTEKNFKWIARSADGWITTPR-------------
MMAR_4546|M.marinum_M ---PVQSHIPVLVGAAGTEKNFKWIARSADGWITTPR-------------
MAV_1077|M.avium_104 ---PVQPHIPVLVGAAGNEKNFKWIARSADGWITTPR-------------
Mflv_1871|M.gilvum_PYR-GCK ---PVQSHIPVLVGAAGTEKNFKWIAKSADGWITTPR-------------
Mvan_4864|M.vanbaalenii_PYR-1 ---PVQAHIPVLVGAAGTEKNFKWIAKSADGWITTPR-------------
MSMEG_5520|M.smegmatis_MC2_155 ---PVQAHIPVLVGAAGNEKNFKWIARSADGWITTPR-------------
TH_2830|M.thermoresistible__bu ---PVQAHIPVLVGAAGTEKNFKWIARSADGWITTPR-------------
MAB_1059c|M.abscessus_ATCC_199 ---TVQKHVPVIVGAGGTEKNFTWIAKNADGWMSTPR-------------
MUL_3458|M.ulcerans_Agy99 ---PVTKPHPPLYIGGGP-ASFGRIARLGAGWIAISP-------------
MLBr_00348|M.leprae_Br4923 ITHPLRADIPIMLGAEGP-KNVALAAEICDGWLPIFFSPRMADMYNEWLD
.: * : . * .. *. **:.
Mb0978c|M.bovis_AF2122/97 -------------DVDIDEPVKLLQDIWAAAGRDGLPQIVALDV-----K
Rv0953c|M.tuberculosis_H37Rv -------------DVDIDEPVKLLQDIWAAAGRDGLPQIVALDV-----K
MMAR_4546|M.marinum_M -------------DFDIDAPVKLLQDTWAAAGRDGAPQIVALDF-----K
MAV_1077|M.avium_104 -------------DFDIDEPVKLLQDTWAAAGRDGAPQIVALDF-----K
Mflv_1871|M.gilvum_PYR-GCK -------------DFTIDEPVKLLQDTWAAAGRDGAPQIVALDF-----K
Mvan_4864|M.vanbaalenii_PYR-1 -------------DFTIDEPVKLLQDTWAAAGRDGAPQIVALDF-----K
MSMEG_5520|M.smegmatis_MC2_155 -------------DFDIDEPVKLLQDIWAAAGREGAPQIRALDY-----K
TH_2830|M.thermoresistible__bu -------------DVDIEEPVKLLQDIWADAGRDGAPQIVALDF-----K
MAB_1059c|M.abscessus_ATCC_199 -------------DEDIAGKIVALKEAWKAAGRDGDPLIYVLAG-----K
MUL_3458|M.ulcerans_Agy99 -------------SPQLLAGP--LEELRSLAG-EEVAVTVCLSD-----E
MLBr_00348|M.leprae_Br4923 EGFARTGARRSREDFEICASAQIVVTEDRAAAFAGIKPFLALYMGGMGAE
. : : *. * :
Mb0978c|M.bovis_AF2122/97 PVPDKLARWAELGVTEVLFGMPDRSADD------------------AAAY
Rv0953c|M.tuberculosis_H37Rv PVPDKLARWAELGVTEVLFGMPDRSADD------------------AAAY
MMAR_4546|M.marinum_M PVPEKLARWAELGVTEVLFGLPDRPEDE------------------IAAY
MAV_1077|M.avium_104 PDAEKLARWAELGVTEVLFGLPDKSEDE------------------VAAY
Mflv_1871|M.gilvum_PYR-GCK PDPDKLAHWRELGVTEVLFGLPDKSESE------------------VAAY
Mvan_4864|M.vanbaalenii_PYR-1 PDPEKLAHWREIGVTEVLFGLPDKSEAD------------------VAAY
MSMEG_5520|M.smegmatis_MC2_155 PDAEKLAHWRDLGVTEVVFGLPDKSEAE------------------VAAY
TH_2830|M.thermoresistible__bu PDPDKLARWRDLGVTEVLFGLPDRSDDE------------------IAAY
MAB_1059c|M.abscessus_ATCC_199 PDPEQLAAWRELGVTEAIFGMPDRSEEE------------------VRAY
MUL_3458|M.ulcerans_Agy99 TKVSDLEGYLPLGVERVLLGLPTEPRDQ------------------TLRY
MLBr_00348|M.leprae_Br4923 GTNFHADVYRRMGYAEVVDEVTALFRSNQKDKAAEIIPNELVDSTMIVGD
. : :* ..: :. :
Mb0978c|M.bovis_AF2122/97 VERLAAKLACCV------------------------
Rv0953c|M.tuberculosis_H37Rv VERLAAKLACCV------------------------
MMAR_4546|M.marinum_M VERLAGKLAAMV------------------------
MAV_1077|M.avium_104 VERLAGKLAALV------------------------
Mflv_1871|M.gilvum_PYR-GCK VERLAGKLAQLV------------------------
Mvan_4864|M.vanbaalenii_PYR-1 VERLAGKLGALT------------------------
MSMEG_5520|M.smegmatis_MC2_155 VERLAGKLSALV------------------------
TH_2830|M.thermoresistible__bu VERLAGKLGLS-------------------------
MAB_1059c|M.abscessus_ATCC_199 IDRLSGKLAAFA------------------------
MUL_3458|M.ulcerans_Agy99 LDQMESELVQLG------------------------
MLBr_00348|M.leprae_Br4923 VDYVCKQIAAWSAAGVTMMMVSASSVEQVRDLAGLF
:: : ::