For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSEMTEDRNHVVVIGGGYAGIMAANHLRSRADLEITVVNPRPVFVERIRLHQLAAGSGTATVDYDSMLGD GIHLVVDAVDRIDADEHRLLLTSGSELRYDYLIYAVGSTAAMPAVAGAAEFAFSVADLDGATRLRHAMAD LPADAAITVVGGGLTGIEAAAELAERGRRVTLVCGGRLGPSLSNRGRRSVARQLGKLGVNVLESAAVSEV RWNAVVLADDTVLASAATVWTAGFGVPDLAARSGLRTDDLGRLLTDETLTSVSDDRIVAAGDAVAPSGQP LRMSCQAAGPLGAQAANTVLSRIAGDQPVALNQAFVGQCISVGRSYGTIQLARTDDTPVNVIIGGRAGAA VKETICKGTLWALRREAAKPGSYFWLKGGSRPVQPSAEQPVVLS
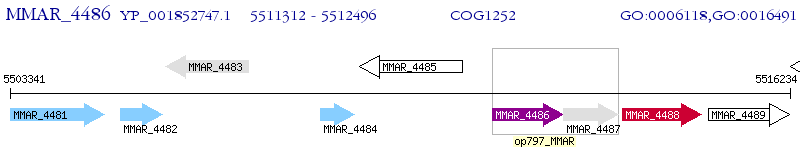
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4486 | - | - | 100% (394) | dehydrogenase |
| M. marinum M | MMAR_2688 | - | 6e-30 | 30.32% (376) | NADH dehydrogenase |
| M. marinum M | MMAR_2728 | ndh | 2e-16 | 28.16% (348) | NADH dehydrogenase Ndh |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1842c | - | 5e-30 | 32.22% (329) | dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1928 | - | 1e-117 | 57.63% (413) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| M. tuberculosis H37Rv | Rv1812c | - | 5e-30 | 32.22% (329) | dehydrogenase |
| M. leprae Br4923 | MLBr_02061 | - | 8e-16 | 27.91% (326) | putative dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1127c | - | 1e-113 | 56.88% (378) | putative oxidoreductase |
| M. avium 104 | MAV_1130 | - | 1e-161 | 74.02% (381) | dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5443 | - | 1e-127 | 59.90% (384) | dehydrogenase |
| M. thermoresistible (build 8) | TH_1141 | - | 1e-131 | 63.28% (384) | dehydrogenase |
| M. ulcerans Agy99 | MUL_4658 | - | 0.0 | 99.24% (393) | dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4805 | - | 1e-134 | 64.87% (390) | FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1842c|M.bovis_AF2122/97 -------------MTRVVVIGSGFAGLWAALGAARRLDELAVPAGTVDVM
Rv1812c|M.tuberculosis_H37Rv -------------MTRVVVIGSGFAGLWAALGAARRLDELAVLAGTVDVM
MMAR_4486|M.marinum_M -----MSEMTEDR-NHVVVIGGGYAGIMAAN-HLRSRADLEITVVNPRPV
MUL_4658|M.ulcerans_Agy99 -----MSEMTEDR-NHVVVIGGGYAGIMAAN-HLRSRADLEITVVNPRPV
MAV_1130|M.avium_104 -----MTEVTRSK-THVVVVGGGYAGTLAAN-HLRQRPDIDITLVNPRPV
TH_1141|M.thermoresistible__bu -----MTSNTGTSVSRVVVIGGGYAGVMAAN-RLRLDPSVDVTLINPRPD
Mvan_4805|M.vanbaalenii_PYR-1 -----MTAQR----THVVVIGGGYAGVMAAN-RLMSRDGLRVTLVNPRPE
Mflv_1928|M.gilvum_PYR-GCK -----MTAQN----IRVIVVGGGYAGVMAAN-RLRSRDDLHVTLVNPRPQ
MSMEG_5443|M.smegmatis_MC2_155 -----MTPNA----PHIVVIGGGYAGVLAAN-HLQQAAHAKITLVNPRPE
MAB_1127c|M.abscessus_ATCC_199 -----MTENT-----TVIVIGGGYAGTLAAN-QLLANKNLTVTLVNPRPH
MLBr_02061|M.leprae_Br4923 MNAQPAATAAQDRRHQVVIIGSGFGGLNAAK--KLKRANVDIKLIARTTH
::::*.*:.* ** :
Mb1842c|M.bovis_AF2122/97 VVSNKPFHDIRVRN-----------------------------YEADLSA
Rv1812c|M.tuberculosis_H37Rv VVSNKPFHDIRVRN-----------------------------YEADLSA
MMAR_4486|M.marinum_M FVERIRLHQLAAGS----------------------------------GT
MUL_4658|M.ulcerans_Agy99 FVERIRLHQLAAGS----------------------------------GT
MAV_1130|M.avium_104 FVERIRLHQLVADT----------------------------------GT
TH_1141|M.thermoresistible__bu FVERIRLHQLVTGS----------------------------------DD
Mvan_4805|M.vanbaalenii_PYR-1 FVERIRLHQFVTGS----------------------------------DD
Mflv_1928|M.gilvum_PYR-GCK FVERIRLHQLAADAGGTAGRAKRRGTSTAGRAKRRGTSTAGRAKRRGTSS
MSMEG_5443|M.smegmatis_MC2_155 FVERIRLHQLAVGN----------------------------------HS
MAB_1127c|M.abscessus_ATCC_199 FVERIRLHQLVAGN----------------------------------AT
MLBr_02061|M.leprae_Br4923 HLFQPLLYQVATGIIS------------------------------EGEI
: . :::. .
Mb1842c|M.bovis_AF2122/97 CRIPLGDVLGPAGVAHVTAEVTAIDADGRRVT---TSTGASYSYDRLVLA
Rv1812c|M.tuberculosis_H37Rv CRIPLGDVLGPAGVAHVTAEVTAIDADGRRVT---TSTGASYSYDRLVLA
MMAR_4486|M.marinum_M ATVDYDSMLGDG-IHLVVDAVDRIDADEHRLL---LTSGSELRYDYLIYA
MUL_4658|M.ulcerans_Agy99 ATVDYDSMLGDG-IHLVVDAVDRIDADEHRLL---LTSGSELGYDYLIYA
MAV_1130|M.avium_104 ATADYATLLGEG-IELIVDTVDHIDATARRAV---LTSGAGLDYDYLIYA
TH_1141|M.thermoresistible__bu AVVDFATVLHPD-VRLVVDTAERIDAADRSVA---LASGGTVSYDHLIYA
Mvan_4805|M.vanbaalenii_PYR-1 AVVDYADVVGEG-IALIVDSVTRIDAPTRRVE---LASGGSIDYDYLIYA
Mflv_1928|M.gilvum_PYR-GCK AVAEFGAVLNPG-VQLVVDNVERIDAPVRRLA---LSSGTSLDYDYLVYA
MSMEG_5443|M.smegmatis_MC2_155 ATTGYDTVLGDR-VRLLVDSVERIDAPIRQVQ---LASGDVVDYDYLLYA
MAB_1127c|M.abscessus_ATCC_199 ATAGYDKLLNPA-VSLVVDKVTYIDAGAQKLE---LASGAVLPYDYLVYA
MLBr_02061|M.leprae_Br4923 APPTRVVLRKQRNIQVLLGNVTHIDLANQCVVSELLGHTYETLYDSLIVA
. : : : . ** : ** *: *
Mb1842c|M.bovis_AF2122/97 SGSHVVKPA-LPGLAEFGFDVDTYDGAVRLQQHLQGLAGGPLTS------
Rv1812c|M.tuberculosis_H37Rv SGSHVVKPA-LPGLAEFGFDVDTYDGAVRLQQHLQGLAGGPLTS------
MMAR_4486|M.marinum_M VGSTAAMPA-VAGAAEFAFSVADLDGATRLRHAMADLP------------
MUL_4658|M.ulcerans_Agy99 VGSTAAMPA-VAGAAEFAFSVADLDGATRLRHAMADLP------------
MAV_1130|M.avium_104 VGSTGAVAE-VPGCAEFAHSVADLESAQRLHYALADLP------------
TH_1141|M.thermoresistible__bu VGSNAGPST-VPGVGEFAYSVTEFEDAQRLRRAVDARP------------
Mvan_4805|M.vanbaalenii_PYR-1 VGSTGAAPT-VPGAAEFAHPISEYEHGAPLRTAVTAAA------------
Mflv_1928|M.gilvum_PYR-GCK VGSTGAVPDGVPGAAEFAYPLAELEQAQRLRSRLADVA------------
MSMEG_5443|M.smegmatis_MC2_155 VGSTGGVPTSVPGAAEFAYPLSELEQAQRLCERLQDIP------------
MAB_1127c|M.abscessus_ATCC_199 VGSTTSTPD-VPGVAEYALSINEFEHAQQVRTRYERLA------------
MLBr_02061|M.leprae_Br4923 AGAGQSYFG-NDQFAEFAPGMKSIDDALELRGRILSAFEQAERSNDPERR
*: .*:. : : . :
Mb1842c|M.bovis_AF2122/97 -AAATVVVVGAGLTGIETACELPGRLHALFA----RGDGVTPRVVLIDHN
Rv1812c|M.tuberculosis_H37Rv -AAATVVVVGAGLTGIETACELPGRLHALFA----RGDGVTPRVVLIDHN
MMAR_4486|M.marinum_M -ADAAITVVGGGLTGIEAAAELAE----------------RGRRVTLVCG
MUL_4658|M.ulcerans_Agy99 -ADAAITVVGGGLTGIEAAAELAE----------------RGRRVTLVCG
MAV_1130|M.avium_104 -LPAPITVVGGGLTGIETASELAE----------------LGRPVTLICG
TH_1141|M.thermoresistible__bu -CDAPITVVGAGPTGLEMAAELAE----------------QGRTVTLVG-
Mvan_4805|M.vanbaalenii_PYR-1 -ADAPIVVVGAGPTGLETAAELAE----------------EGRRVTLVCG
Mflv_1928|M.gilvum_PYR-GCK -AIAPVVVIGGGLTGIEAAAEFAE----------------AGRDVTLVS-
MSMEG_5443|M.smegmatis_MC2_155 -MSAPVVVVGGGLTGIEAAGEFAE----------------AGRPVTLIT-
MAB_1127c|M.abscessus_ATCC_199 -PDAPIVVVGAGLTGVELAGELAE----------------AGRRVTLICG
MLBr_02061|M.leprae_Br4923 EKLLTFTVVGAGPTGVEMAGQIAELADHTLKGAFRRIDSTKARVVLLDAA
...*:*.* **:* * ::. * * :
Mb1842c|M.bovis_AF2122/97 PFVGSDMGLSARPVIEQALLDNGVETRTG--VSVAAVSPGGVTLSSG---
Rv1812c|M.tuberculosis_H37Rv PFVGSDMGLSARPVIEQALLDNGVETRTG--VSVAAVSPGGVTLSSG---
MMAR_4486|M.marinum_M GRLGPSLSNRGRRSVARQLGKLGVNVLES--AAVSEVRWNAVVLADD---
MUL_4658|M.ulcerans_Agy99 GRLGPSLSNRGRRSVARQLGKLGVNVLES--AAVSEVRWNAVVLADD---
MAV_1130|M.avium_104 PVLGPSLSPRGRRSVAKRLRRLGVGVLES--VTVTEVRWNEVVLSDG---
TH_1141|M.thermoresistible__bu DAIGPYLSEPGWRAVAKRFRRLGVRTVE---ARVAEVRADAVLLADG---
Mvan_4805|M.vanbaalenii_PYR-1 SVLGPYLSTRGRRAAAKRMTKLGVDVIDGPGATVTEVLADAVVLADG---
Mflv_1928|M.gilvum_PYR-GCK PVVGPSLSGPGRRSVTRRLRKLGVHLVSAS---VRAVGADHVVLDDA---
MSMEG_5443|M.smegmatis_MC2_155 DVVGASLGDQARRSVVKALRKLGVTIVSGPDILVASVGATDVTLADG---
MAB_1127c|M.abscessus_ATCC_199 TQLLPSVGEPARRAAAKRLRKLGVDVQAP--ATAIRVDENSVTLSDG---
MLBr_02061|M.leprae_Br4923 PAVLPPMGEKLGQRAAARLQKMGVEIQLS--AMVTDVDRNGITVQDSDGT
: . :. : ** . * : : .
Mb1842c|M.bovis_AF2122/97 -ERLAAATVVWCAGMRAS----RLTEQLPVARDRLGRLQVDDYLRVIGVP
Rv1812c|M.tuberculosis_H37Rv -ERLAAATVVWCAGMRAS----RLTEQLPVARDRLGRLQVDDYLRVIGVP
MMAR_4486|M.marinum_M -TVLASAATVWTAGFGVP----DLAARSGLRTDDLGRLLTDETLTSVSDD
MUL_4658|M.ulcerans_Agy99 -TVLASAATVWTAGFGVP----DLAARSGLRTDDLGRLLTDETLTSVSDD
MAV_1130|M.avium_104 -VVLPSAATVWTAGFSVP----DLAARSGLRTDPLGRLLTDETLTSIDDD
TH_1141|M.thermoresistible__bu -RRLDSAVTVWTAGFAVP----DLAARSGLSTDAIGRLRTDETLTSVDDD
Mvan_4805|M.vanbaalenii_PYR-1 -RRVASAVTIWTAGFGVP----DLAVRSGLSTDAIGRLLTDETLASVDDP
Mflv_1928|M.gilvum_PYR-GCK -RSLPSAVTVWTAGFGVP----TLAADSGLATDPLGRLLTDETLTSVDDD
MSMEG_5443|M.smegmatis_MC2_155 -TRLPSAVTVWTTGFGVP----GLAAASGLTTDELGRLITDETLTSVDDD
MAB_1127c|M.abscessus_ATCC_199 -RVLPSALTVWTAGFGVP----RLAIDSGLRTDALGRLLTDETLVSLDNP
MLBr_02061|M.leprae_Br4923 VRRIESACKVWSAGVSASRLGRDLAEQSLVELDWAGRVKVLPDLSIPGYR
: :* :* :*. .. *: : * **: . * .
Mb1842c|M.bovis_AF2122/97 AMFAAGDVAAARMDDEHLSVMSCQHGRPMGRYAGCNVINDLFDQPLLALR
Rv1812c|M.tuberculosis_H37Rv AMFAAGDVAAARMDDEHLSVMSCQHGRPMGRYAGCNVINDLFDQPLLALR
MMAR_4486|M.marinum_M RIVAAGDAVAP---SGQPLRMSCQAAGPLGAQAANTVLSRIAGDQPVALN
MUL_4658|M.ulcerans_Agy99 RIVAAGDAVAP---SGQPLRMSCQAAGPLGAQAANTVLSRIAGDQPVALN
MAV_1130|M.avium_104 RIVAAGDAAAP---SGRPLRMSCQAAGPLGAQAANTVLGRIAGTEPAPLN
TH_1141|M.thermoresistible__bu RIVAAGDAAAP---SGQPLRMSCQAAGPLGAQAADTVLSRIAGDRPAPLD
Mvan_4805|M.vanbaalenii_PYR-1 RIVAAGDAAAP---SDLPLRMSCQAAIPLGAQAANTVLSRIAGTTPKPIN
Mflv_1928|M.gilvum_PYR-GCK RIVAAGDSAAP---SGLPYRMSCQAGLPLGAQAAETVLARIAGTDAAVAT
MSMEG_5443|M.smegmatis_MC2_155 RIVAAGDCASP---SGIPLRMSCQAAGPMGIQAANTVLARLAGTEPAVLN
MAB_1127c|M.abscessus_ATCC_199 RIIAAGDAASP---SGVPLRMSCQAAGPLAARAVKTVLALVDGLRPEART
MLBr_02061|M.leprae_Br4923 NVFVVGDMAAID--GVPGVAQGAIQGAKYVANNIKAELGEANPAEREPFQ
:...** .: . .. . :
Mb1842c|M.bovis_AF2122/97 IPWYVTVLDLGSAGAVYTEG--------------------------WERK
Rv1812c|M.tuberculosis_H37Rv IPWYVTVLDLGSAGAVYTEG--------------------------WERK
MMAR_4486|M.marinum_M QAFVGQCISVGRSYGTIQLARTDDT---------PVNVIIGGRAGAAVKE
MUL_4658|M.ulcerans_Agy99 QAFVGQCISVGRSYGTIQLAHTDDT---------PVNVIIGGRAGAAVKE
MAV_1130|M.avium_104 QAFVGQCISLGRAAATFQLARTDDT---------PINAYLGGRAAAKLKE
TH_1141|M.thermoresistible__bu QAFAGQCISLGRDAGIIQLARRDDT---------AIPVFLSGRAAAWVKE
Mvan_4805|M.vanbaalenii_PYR-1 QAFTGQCISLGRSAGLIQIAHLDDR---------VMPLYIGGRTAASVKE
Mflv_1928|M.gilvum_PYR-GCK VPMSGQCISLGRDAGTVQLQRKDDT---------PLNLYLGGRTGAFVKE
MSMEG_5443|M.smegmatis_MC2_155 QAFTGQCVSVGRTYGTVQIAHSDDT---------PRRLYFGGRTGAAIKE
MAB_1127c|M.abscessus_ATCC_199 QIFTGQNVSIGRRAAVIQVGHTDDT---------PRTLYIGGRTGAAIKE
MLBr_02061|M.leprae_Br4923 YFDKGSMATVSRFSAVAKIGPLEFSGFIAWLMWLVLHLMYLIGFKTKITT
:. .
Mb1842c|M.bovis_AF2122/97 VVSQGAPAKTTKQSINTRRIYPPLNGSRADLLAAAAPRVQPRP-------
Rv1812c|M.tuberculosis_H37Rv VVSQGAPAKTTKQSINTRRIYPPLNGSRADLLAAAAPRVQPRP-------
MMAR_4486|M.marinum_M TICKGTLWALRREAAKPGSYFWLKGGSRPVQPSAEQPVVLS---------
MUL_4658|M.ulcerans_Agy99 AICKGTLWALRREAAKPGSYFWLKGGSRPVQPSAEQPVVLP---------
MAV_1130|M.avium_104 AICRGTLWAIRREAAKPGSYRWLKGGQR--VPADERVGV-----------
TH_1141|M.thermoresistible__bu RVCRGTIWALAREARRPGSYLWLKGGRRPAQPALDEMAVS----------
Mvan_4805|M.vanbaalenii_PYR-1 AVCKGTISFLSREARKPGSYFWLKGGKR--QQKLAAQPVSS---------
Mflv_1928|M.gilvum_PYR-GCK QVCRFTITWLAAEARKPGSYRSLKGPDRSELIASTAVPVR----------
MSMEG_5443|M.smegmatis_MC2_155 QVCKATITFMAKEGRKPGSYFWFKGRNRARQLAEAAKDTVGAARR-----
MAB_1127c|M.abscessus_ATCC_199 MVCKGTLSILATAAKHPTWYYWPQGGKRAEALRAQV--------------
MLBr_02061|M.leprae_Br4923 LLSWTVTFLSTRRGQLTITEQQAFARTRLEQLAELTAESQSTAAANRATM
:. . . . *
Mb1842c|M.bovis_AF2122/97 ---
Rv1812c|M.tuberculosis_H37Rv ---
MMAR_4486|M.marinum_M ---
MUL_4658|M.ulcerans_Agy99 ---
MAV_1130|M.avium_104 ---
TH_1141|M.thermoresistible__bu ---
Mvan_4805|M.vanbaalenii_PYR-1 ---
Mflv_1928|M.gilvum_PYR-GCK ---
MSMEG_5443|M.smegmatis_MC2_155 ---
MAB_1127c|M.abscessus_ATCC_199 ---
MLBr_02061|M.leprae_Br4923 RAS