For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QLGMIGLGRMGANIVRRVVNGGHECVVYDHNPDAVKAMAGENNTTGVSSLSELRDKLSAPRVIWVMVPAG TITTGVIEELATTLDAGDIVIDGGNTYYRDDIKHAKLLSGKGIHMLDCGTSGGVWGLDRGYCLMVGGEPD AFAHAEPILATVAPGVQAAPRTPGRDGEVAEAEKGYLHCGPTGAGHFVKMVHNGIEYGMMASLAEGLNIL RNADVGTRVQQGDAETAPLANPEYYQYDIDIPEVAEVWRRGSVIGSWLLDLTAIALHESPELQEFSGRVS DSGEGRWTSIAAIDEGVPTPVLTTALQSRFASRDLDDFANKALSAMRKQFGGHAEKPVN*
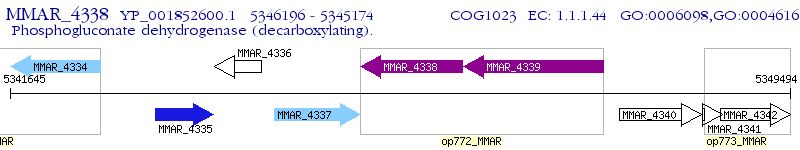
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4338 | gnd2 | - | 100% (340) | 6-phosphogluconate dehydrogenase, decarboxylating Gnd2 |
| M. marinum M | MMAR_2718 | gnd1 | 4e-39 | 31.68% (322) | 6-phosphogluconate dehydrogenase Gnd1 |
| M. marinum M | MMAR_1077 | mmsB | 2e-14 | 25.35% (213) | 3-hydroxyisobutyrate dehydrogenase, MmsB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1153 | gnd2 | 1e-175 | 86.47% (340) | 6-phosphogluconate dehydrogenase-like protein |
| M. gilvum PYR-GCK | Mflv_0825 | - | 3e-74 | 45.00% (340) | 6-phosphogluconate dehydrogenase-like protein |
| M. tuberculosis H37Rv | Rv1122 | gnd2 | 1e-175 | 86.47% (340) | 6-phosphogluconate dehydrogenase-like protein |
| M. leprae Br4923 | MLBr_02065 | gnd | 7e-38 | 30.75% (322) | 6-phosphogluconate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0003 | gnD | 1e-75 | 45.72% (339) | 6-phosphogluconate dehydrogenase-like protein |
| M. avium 104 | MAV_1253 | gnd | 1e-178 | 87.65% (340) | 6-phosphogluconate dehydrogenase-like protein |
| M. smegmatis MC2 155 | MSMEG_0002 | - | 1e-75 | 45.88% (340) | 6-phosphogluconate dehydrogenase-like protein |
| M. thermoresistible (build 8) | TH_3444 | - | 1e-111 | 57.57% (337) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0148 | gnd2 | 0.0 | 99.71% (340) | 6-phosphogluconate dehydrogenase-like protein |
| M. vanbaalenii PYR-1 | Mvan_0003 | - | 5e-74 | 45.00% (340) | 6-phosphogluconate dehydrogenase-like protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0825|M.gilvum_PYR-GCK ----------MQLGLIGLGKMGFNMRARLRDGGHDVVGYD----PRPE--
Mvan_0003|M.vanbaalenii_PYR-1 ----------MQLGLIGLGKMGFNMRARLRDGGHDVVGFD----PRPE--
MSMEG_0002|M.smegmatis_MC2_155 ----------MQLGLIGLGKMGFNMRERLREAGHEVIGYD----PRPE--
MAB_0003|M.abscessus_ATCC_1997 ----------MQLGLVGLGKMGFNMRDRLRAGGHEVIGYD----PRPE--
Mb1153|M.bovis_AF2122/97 ----------MQLGMIGLGRMGANIVRRLAKGGHDCVVYD----HDPDAV
Rv1122|M.tuberculosis_H37Rv ----------MQLGMIGLGRMGANIVRRLAKGGHDCVVYD----HDPDAV
MMAR_4338|M.marinum_M ----------MQLGMIGLGRMGANIVRRVVNGGHECVVYD----HNPDAV
MUL_0148|M.ulcerans_Agy99 ----------MQLGMIGLGRMGADIVRRVVNGGHECVVYD----HNPDAV
MAV_1253|M.avium_104 ----------MQLGMIGLGRMGANIVRRVVKGGHECVVYD----HNPDAV
TH_3444|M.thermoresistible__bu ----------MQLGMIGAGRMGSGLVRRLTAREHSCVVYD----PDPAAV
MLBr_02065|M.leprae_Br4923 MSAPESKTAIAQIGVTGLAVMGSNIARNFARHGYTVALHNRSIAKTDTLL
*:*: * . ** .: .. : .:
Mflv_0825|M.gilvum_PYR-GCK --------VSDVESLEELAQRLDAPRVVWVMVPSGTVTDSTITELAEVLS
Mvan_0003|M.vanbaalenii_PYR-1 --------VSDVASLADLAARLSAPRVVWVMVPSGTVTDSTISELAGVLS
MSMEG_0002|M.smegmatis_MC2_155 --------VSDVANLEELAGALSAPRVVWVMVPSGTITHSTIVELAGVLS
MAB_0003|M.abscessus_ATCC_1997 --------VTDVPSLKGLAEALDAPRVVWVMVPSGPITQQTIAELSEELS
Mb1153|M.bovis_AF2122/97 KAMAGEDRTTGVASLRELSQRLSAPRVVWVMVPAGNITTAVIEELANTLE
Rv1122|M.tuberculosis_H37Rv KAMAGEDRTTGVASLRELSQRLSAPRVVWVMVPAGNITTAVIEELANTLE
MMAR_4338|M.marinum_M KAMAGENNTTGVSSLSELRDKLSAPRVIWVMVPAGTITTGVIEELATTLD
MUL_0148|M.ulcerans_Agy99 KAMAGENNTTGVSSLSELRDKLSAPRVIWVMVPAGTITTGVIEELATTLD
MAV_1253|M.avium_104 KAMAGEEKTTAVSSLQELAAKLSAPRVVWVMVPAGTITTGVIEELATTLE
TH_3444|M.thermoresistible__bu GRLP-DTHVSRAESFAELVQALAIPRIVWVMVPAAR-TGEVIAELEGLME
MLBr_02065|M.leprae_Br4923 KEHGSEGNFVRTETIPEFLAALQTPRRVLIMVKAGDATDAVINELADVME
. .: : * ** : :** :. * .* ** :.
Mflv_0825|M.gilvum_PYR-GCK EGDLVIDGGNSRYTEDQPHAKMLAQKGIGFVDAGVSGGIWGLTEGYGLMV
Mvan_0003|M.vanbaalenii_PYR-1 PGDLVIDGGNSRYTEDGRHANLLAEKGIAFIDAGVSGGIWGLTEGYGLMV
MSMEG_0002|M.smegmatis_MC2_155 PGDLVIDGGNSRYTEDGPHAELLGEKGISFIDAGVSGGIWGLTEGYGLMV
MAB_0003|M.abscessus_ATCC_1997 EGDLVIDGGNSRFTEDKPNADLLAAKGIGYIDAGVSGGVWGKENGYGLMV
Mb1153|M.bovis_AF2122/97 AGDIVIDGGNTYYRDDLRHEKLLFKKGIHLLDCGTSGGVWGRERGYCLMI
Rv1122|M.tuberculosis_H37Rv AGDIVIDGGNTYYRDDLRHEKLLFKKGIHLLDCGTSGGVWGRERGYCLMI
MMAR_4338|M.marinum_M AGDIVIDGGNTYYRDDIKHAKLLSGKGIHMLDCGTSGGVWGLDRGYCLMV
MUL_0148|M.ulcerans_Agy99 AGDIVIDGGNTYYRDDIKHAKLLSGKGIHMLDCGTSGGVWGLDRGYCLMV
MAV_1253|M.avium_104 PGDIVIDGGNSYYRDDIRHSKTLSEKGIRLLDCGTSGGVWGRERGYCLMI
TH_3444|M.thermoresistible__bu FDDVIVDGGNSYYRDAIARSRRLASRGIHYLDVGTSGGVHGADRGFCLMI
MLBr_02065|M.leprae_Br4923 PSDIIIDGGNSLFTDTIRREKAMRERGLHFVGAGISGGEEGALNGPSIMP
.*:::****: : : . : :*: :. * *** * .* :*
Mflv_0825|M.gilvum_PYR-GCK GGSDADIARAMPIFDTLRP--------PG------PQEDGFVHVGPVGAG
Mvan_0003|M.vanbaalenii_PYR-1 GGSDADVARAMPIFETLRP--------PG------PLEDGFVHVGPVGAG
MSMEG_0002|M.smegmatis_MC2_155 GGSDDDVARAMPIFDALRP--------PG------DVADGFVHAGPVGAG
MAB_0003|M.abscessus_ATCC_1997 GGSDEDVARAMPIFDTLRP--------EG------ERADGFVHAGPVGAG
Mb1153|M.bovis_AF2122/97 GGDGDAFARAEPIFATVAPGVAAAPRTPGRDGEVAPSEQGYLHCGPCGSG
Rv1122|M.tuberculosis_H37Rv GGDGDAFARAEPIFATVAPGVAAAPRTPGRDGEVAPSEQGYLHCGPCGSG
MMAR_4338|M.marinum_M GGEPDAFAHAEPILATVAPGVQAAPRTPGRDGEVAEAEKGYLHCGPTGAG
MUL_0148|M.ulcerans_Agy99 GGEPDAFAHAEPILATVAPGVQAAPRTPGRDGEVAEAEKGYLHCGPTGAG
MAV_1253|M.avium_104 GGDEEAFAHAEPIFATVAPGVDAAPRTPGRDGEVTRSEQGYLHCGPSGAG
TH_3444|M.thermoresistible__bu GGESRIVERLEPIFRALAPGADTATRTPGRTGGITPAEHGYLHCGPAGSG
MLBr_02065|M.leprae_Br4923 GGPAESYTSLGPLLEEISAHVDG--------------VSCCTHIGPGGSG
** *:: : . * ** *:*
Mflv_0825|M.gilvum_PYR-GCK HFAKMVHNGIEYALMTAYAEGYELLAAEELVKDPQ---------------
Mvan_0003|M.vanbaalenii_PYR-1 HFAKMVHNGIEYALMTAYAEGYEMLAAEDLVKDPQ---------------
MSMEG_0002|M.smegmatis_MC2_155 HYAKMVHNGIEYGLMHAYAEGYELLAAEELIKDPQ---------------
MAB_0003|M.abscessus_ATCC_1997 HYAKMIHNGIEYGLMHAYAEGYELLAAEPLITDVQ---------------
Mb1153|M.bovis_AF2122/97 HFVKMVHNGIEYGMMASLAEGLNILRNADVGTRVQHGDAETAPLPNPECY
Rv1122|M.tuberculosis_H37Rv HFVKMVHNGIEYGMMASLAEGLNILRNADVGTRVQHGDAETAPLPNPECY
MMAR_4338|M.marinum_M HFVKMVHNGIEYGMMASLAEGLNILRNADVGTRVQQGDAETAPLANPEYY
MUL_0148|M.ulcerans_Agy99 HFVKMVHNGIEYGMMASLAEGLNILRNADVGTRVQQGDAETAPLANPEYY
MAV_1253|M.avium_104 HFVKMVHNGIEYGMMASLAEGLNVLRNADIGKHVQEGDAETAPLSNPEFY
TH_3444|M.thermoresistible__bu HFVKMVHNGVEYGQMAALAEGMAILRNADVGAETRSVDAETAPLADADCY
MLBr_02065|M.leprae_Br4923 HFVKMVHNGIEYSDMQLIGEAYQLLRDGLGMSAPQ---------------
*:.**:***:**. * .*. :* :
Mflv_0825|M.gilvum_PYR-GCK -------AVYQAWTNGTVVRSWLQQLLAKALKED------PDLSEISGYT
Mvan_0003|M.vanbaalenii_PYR-1 -------AVYQAWTNGTVVRSWLQQLLAKALKED------PSLSDISGYT
MSMEG_0002|M.smegmatis_MC2_155 -------AVIQAWSNGTVVRSWLQSLLAKALKED------PGFAEISGYT
MAB_0003|M.abscessus_ATCC_1997 -------AVLQAWTKGTVVRSWLLDLLAKALKED------PGFDAISGYT
Mb1153|M.bovis_AF2122/97 QYDFDIPEVAEVWRRGSVIGSWLLDLTAIALRES------PDLAEFSGRV
Rv1122|M.tuberculosis_H37Rv QYDFDIPEVAEVWRRGSVIGSWLLDLTAIALRES------PDLAEFSGRV
MMAR_4338|M.marinum_M QYDIDIPEVAEVWRRGSVIGSWLLDLTAIALHES------PELQEFSGRV
MUL_0148|M.ulcerans_Agy99 QYDIDIPEVAEVWRRGSVIGSWLLDLTAIALHES------PELQEFSGRV
MAV_1253|M.avium_104 QYDFDIPEVAEVWRRGSVIGSWLLDLTAIALHES------PELKEFSGRV
TH_3444|M.thermoresistible__bu TYDFDIADILEVWRRGSVVSSWLVDLTAAAVYAN------PQLDEFSAEV
MLBr_02065|M.leprae_Br4923 -----IADVFTEWNRGDLN-SYLVEITAEVLRQTDIKTGRPLVDVILDKA
: * .* : *:* .: * .: * . : .
Mflv_0825|M.gilvum_PYR-GCK EDSGEGRWTVEEAIRLRVPVPGIAASLFARFLS-----------------
Mvan_0003|M.vanbaalenii_PYR-1 EDSGEGRWTVEEAIRLRVPVPGIAASLFARFLS-----------------
MSMEG_0002|M.smegmatis_MC2_155 EDSGEGRWTVEEAIAHRVPMPVIAASLFARFAS-----------------
MAB_0003|M.abscessus_ATCC_1997 EDSGEGRWTVEEAINHRVPAPVIAASLFARFAS-----------------
Mb1153|M.bovis_AF2122/97 SDSGEGRWTAIAAIDEGVPAPVLTTALQSRFAS-----------------
Rv1122|M.tuberculosis_H37Rv SDSGEGRWTAIAAIDEGVPAPVLTTALQSRFAS-----------------
MMAR_4338|M.marinum_M SDSGEGRWTSIAAIDEGVPTPVLTTALQSRFAS-----------------
MUL_0148|M.ulcerans_Agy99 SDSGEGRWTSIAAIDEGVPTPVLTTALQSRFAS-----------------
MAV_1253|M.avium_104 SDSGEGRWTSIAAIDEGVPTPVLTTALQSRFAS-----------------
TH_3444|M.thermoresistible__bu DDSGEGRWAVQTAVDEGVPAPVLTAALYERFAS-----------------
MLBr_02065|M.leprae_Br4923 EQKGTGRWTVQSALDLGVPITGIAEAVFARALSGSVLQRKAAIGLASGKL
.:.* ***: *: ** . :: :: * *
Mflv_0825|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0003|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0002|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0003|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1153|M.bovis_AF2122/97 --------------------------------------------------
Rv1122|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4338|M.marinum_M --------------------------------------------------
MUL_0148|M.ulcerans_Agy99 --------------------------------------------------
MAV_1253|M.avium_104 --------------------------------------------------
TH_3444|M.thermoresistible__bu --------------------------------------------------
MLBr_02065|M.leprae_Br4923 GNKPTDRETFIEDVRQALYASKIVAYAQGFNHIQTGSTEFGWNITPGDLA
Mflv_0825|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0003|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0002|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0003|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1153|M.bovis_AF2122/97 --------------------------------------------------
Rv1122|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4338|M.marinum_M --------------------------------------------------
MUL_0148|M.ulcerans_Agy99 --------------------------------------------------
MAV_1253|M.avium_104 --------------------------------------------------
TH_3444|M.thermoresistible__bu --------------------------------------------------
MLBr_02065|M.leprae_Br4923 TIWRGGCIIRAKFLNRIKEAFDAAPDLASLIVAPYFRSAVESAIDSWRRV
Mflv_0825|M.gilvum_PYR-GCK -----------------------RQDESPTMRAVAALRNQFGGHAVKRVS
Mvan_0003|M.vanbaalenii_PYR-1 -----------------------RQDESPTMKAVAALRNQFGGHAVKRVS
MSMEG_0002|M.smegmatis_MC2_155 -----------------------RQEDSPTMKAVSALRNQFGGHAVKRIS
MAB_0003|M.abscessus_ATCC_1997 -----------------------RQEDSPAMKAVSALRNQFGGHAVKRVG
Mb1153|M.bovis_AF2122/97 -----------------------RDLDDFANKALSAMRKQFGGHAEKPAN
Rv1122|M.tuberculosis_H37Rv -----------------------RDLDDFANKALSAMRKQFGGHAEKPAN
MMAR_4338|M.marinum_M -----------------------RDLDDFANKALSAMRKQFGGHAEKPVN
MUL_0148|M.ulcerans_Agy99 -----------------------RDLDDFANKALSAMRKQFGGHAEKPVN
MAV_1253|M.avium_104 -----------------------RHLDDFANKTLSAMRKQFGGHAEKPAN
TH_3444|M.thermoresistible__bu -----------------------RGEADYSRKALSAMRHAFGGHRRKGAA
MLBr_02065|M.leprae_Br4923 VSTATQLGIPNPGFSSALSYYDALRTERLPAALTQAQRDFFGAHSYGRVD
. * *. **.*
Mflv_0825|M.gilvum_PYR-GCK ESG---------------
Mvan_0003|M.vanbaalenii_PYR-1 ESG---------------
MSMEG_0002|M.smegmatis_MC2_155 ESG---------------
MAB_0003|M.abscessus_ATCC_1997 LSG---------------
Mb1153|M.bovis_AF2122/97 ------------------
Rv1122|M.tuberculosis_H37Rv ------------------
MMAR_4338|M.marinum_M ------------------
MUL_0148|M.ulcerans_Agy99 ------------------
MAV_1253|M.avium_104 ------------------
TH_3444|M.thermoresistible__bu R-----------------
MLBr_02065|M.leprae_Br4923 ARGKFHTLWSEDRSEVTV