For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSSESSTATAQIGVTGLAVMGSNIARNFARHGYTVALHNRSIAKTDALLKEHGAEGKFVRSETISEFLAA LETPRRVLIMVKAGEPTDAVINELADAMEPGDIIIDGGNALYTDTIRREKAVRERGLHFVGAGISGGEEG ALNGPSIMPGGPAESYTSLGPLLEEISAHVDGVPCCTHIGPDGSGHFVKMVHNGIEYSDMQLIGEAYQLL RDGLGMSAPQIADVFSEWNKGDLDSYLVEITAEVLRQTDAKTGKPLVDVILDQAEQKGTGRWTVKSALDL GVPVTGIAEAVFARALSGSVQQRAAAVGLPSGRLGDKPADPDTFTENVRRALYASKIVAYAQGFNQIQAG SAEFGWDITPGDLATIWRGGCIIRAKFLNRIKEAFDAEPALASLLVAPYFRSAIESAVDSWRRVVSTAAQ LGIPTPGFSSALAYYDALRTERLPAALTQAQRDFFGAHTYGRIDAAGKFHTLWSADRSEVPA*
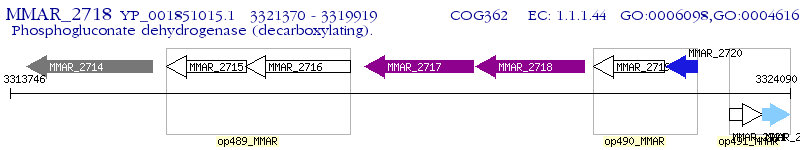
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2718 | gnd1 | - | 100% (483) | 6-phosphogluconate dehydrogenase Gnd1 |
| M. marinum M | MMAR_4338 | gnd2 | 6e-39 | 31.68% (322) | 6-phosphogluconate dehydrogenase, decarboxylating Gnd2 |
| M. marinum M | MMAR_1077 | mmsB | 1e-10 | 25.34% (221) | 3-hydroxyisobutyrate dehydrogenase, MmsB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1875c | gnd1 | 0.0 | 91.12% (383) | 6-phosphogluconate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3359 | - | 0.0 | 83.64% (483) | 6-phosphogluconate dehydrogenase |
| M. tuberculosis H37Rv | Rv1844c | gnd1 | 0.0 | 90.08% (484) | 6-phosphogluconate dehydrogenase |
| M. leprae Br4923 | MLBr_02065 | gnd | 0.0 | 88.57% (481) | 6-phosphogluconate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2413c | - | 0.0 | 84.31% (478) | 6-phosphogluconate dehydrogenase |
| M. avium 104 | MAV_2871 | gnd | 0.0 | 86.19% (485) | 6-phosphogluconate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3632 | gnd | 0.0 | 85.33% (484) | 6-phosphogluconate dehydrogenase |
| M. thermoresistible (build 8) | TH_2383 | gnd | 0.0 | 83.23% (483) | PROBABLE 6-PHOSPHOGLUCONATE DEHYDROGENASE GND1 |
| M. ulcerans Agy99 | MUL_3035 | gnd1 | 0.0 | 99.59% (483) | 6-phosphogluconate dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_3089 | - | 0.0 | 85.07% (489) | 6-phosphogluconate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2718|M.marinum_M -----MSSSE----SSTATAQIGVTGLAVMGSNIARNFARHGYTVALHNR
MUL_3035|M.ulcerans_Agy99 -----MSSSE----SSTATAQIGVTGLAVMGSNIARNFARHGYTVALHNR
MLBr_02065|M.leprae_Br4923 -----MSAPE----SKTAIAQIGVTGLAVMGSNIARNFARHGYTVALHNR
Mflv_3359|M.gilvum_PYR-GCK -----MSSS-----GTTGSAQIGVTGLAVMGSNIARNFARHGYTVALHNR
Mvan_3089|M.vanbaalenii_PYR-1 -----MSSSQ----STTGTAQIGVTGLAVMGSNIARNFARHGYTVALHNR
MAV_2871|M.avium_104 MRGEDMSSSVTPSRPTTGTAQIGVTGLAVMGSNIARNFARHGYTVALHNR
TH_2383|M.thermoresistible__bu -----MSTS-----DSTATAQIGVTGLAVMGSNIARNFARNGYTVAVHNR
MSMEG_3632|M.smegmatis_MC2_155 -----MTASSTPS-STTGTAQIGVTGLAVMGSNIARNFARHGYTVALHNR
MAB_2413c|M.abscessus_ATCC_199 -----MTAADG---SAQGKAQIGVTGLAVMGSNIARNFARHGYTVALHNR
Rv1844c|M.tuberculosis_H37Rv -----MSSSE----SPAGIAQIGVTGLAVMGSNIARNFARHGYTVAVHNR
Mb1875c|M.bovis_AF2122/97 -----------------------------MGSNIARNFARHGYTVAVHNR
***********:*****:***
MMAR_2718|M.marinum_M SIAKTDALLKEHGAEGKFVRSETISEFLAALETPRRVLIMVKAGEPT--D
MUL_3035|M.ulcerans_Agy99 SIAKTDALLKEHGAEGKFVRSETISEFLAALETPRRVLIMVKAGEPT--D
MLBr_02065|M.leprae_Br4923 SIAKTDTLLKEHGSEGNFVRTETIPEFLAALQTPRRVLIMVKAGDAT--D
Mflv_3359|M.gilvum_PYR-GCK SVAKTDALLAEHGSDGNFVRSETIAEFLDALEKPRRVLIMVKAGDPT--D
Mvan_3089|M.vanbaalenii_PYR-1 SIAKTDALLAEHGTEGKFVRSETIAEFLDALEKPRRVLIMVKAGDPT--D
MAV_2871|M.avium_104 SIAKTDALLKEHGDEGEFVRCETIAEFLDALEKPRRVLIMVKAGDPT--D
TH_2383|M.thermoresistible__bu TVAKTDALIEQFGSEGDFVRTETIPEFVAALERPRRVLIMVKAGAPT--D
MSMEG_3632|M.smegmatis_MC2_155 SIAKTDALLAEHGSEGKFVRSETIAEFIDALEKPRKVLIMVKAGEAT--D
MAB_2413c|M.abscessus_ATCC_199 SIAKTDALLAEHGSEGNFVRSESIEEFVAALERPRRVLIMVKAGDPT--D
Rv1844c|M.tuberculosis_H37Rv SVAKTDALLKEHSSDGKFVRSETIPEFLAALEKPRRVLIMVKAGEATDAD
Mb1875c|M.bovis_AF2122/97 SVAKTDALLKEHSSDGKFVRSETIPEFLAALEKPRRVLIMVKAGEAT--D
::****:*: :.. :*.*** *:* **: **: **:******** .* *
MMAR_2718|M.marinum_M AVINELADAMEPGDIIIDGGNALYTDTIRREKAVRERGLHFVGAGISGGE
MUL_3035|M.ulcerans_Agy99 AVINELADAMEPGDIIIDGGNALYTDTIRREKAVRERGLHFVGAGISGGE
MLBr_02065|M.leprae_Br4923 AVINELADVMEPSDIIIDGGNSLFTDTIRREKAMRERGLHFVGAGISGGE
Mflv_3359|M.gilvum_PYR-GCK AVINELADAMEEGDIIIDGGNALYTDTIRREKAIRERGLHFVGAGISGGE
Mvan_3089|M.vanbaalenii_PYR-1 AVINELADAMEEGDIIIDGGNALYTDTIRREKAIRERGLHFVGAGISGGE
MAV_2871|M.avium_104 AVINELADAMEPGDIIIDGGNALYTDTIRREQAMRERGLHFVGAGISGGE
TH_2383|M.thermoresistible__bu AVIAELADAMEPGDIIIDGGNSLYTDTIRREKAMAERGLHFVGAGISGGE
MSMEG_3632|M.smegmatis_MC2_155 AVIDELADAMEPGDIIIDGGNALYTDTIRREAAIRARGLHFVGAGISGGE
MAB_2413c|M.abscessus_ATCC_199 AVINELADAMEQGDIIIDGGNSLYTDTIRREKAMADRGLHFVGAGISGGE
Rv1844c|M.tuberculosis_H37Rv AVINELADAMEPGDIIIDGGNALYTDTMRREKAMRERGLHFVGAGISGGE
Mb1875c|M.bovis_AF2122/97 AVINELADAMEPGDIIIDGGNALYTDTMRREKAMRERGLHFVGAGISGGE
*** ****.** .********:*:***:*** *: **************
MMAR_2718|M.marinum_M EGALNGPSIMPGGPAESYTSLGPLLEEISAHVDGVPCCTHIGPDGSGHFV
MUL_3035|M.ulcerans_Agy99 EGALNGPSIMPGGPAESYTSLGPLLEEISAHVDGVPCCTHIGPDGSGHFV
MLBr_02065|M.leprae_Br4923 EGALNGPSIMPGGPAESYTSLGPLLEEISAHVDGVSCCTHIGPGGSGHFV
Mflv_3359|M.gilvum_PYR-GCK EGALNGPSIMPGGPAESYASLGPLLEEISAHVDGVPCCTHIGPDGAGHFV
Mvan_3089|M.vanbaalenii_PYR-1 EGALNGPSIMPGGPKESYESLGPLLEEISAHVDGVPCCTHIGPDGAGHFV
MAV_2871|M.avium_104 EGALNGPSIMPGGPAESYRSLGPLLEEISAHVDGVPCCTHIGPDGAGHFV
TH_2383|M.thermoresistible__bu EGALHGPSIMPGGSAESYRTLGPMLEKISAHVDGVPCCTHIGPDGAGHFV
MSMEG_3632|M.smegmatis_MC2_155 EGALNGPSIMPGGPAESYESLGPLLEEISAHVDGVPCCTHIGPDGAGHFV
MAB_2413c|M.abscessus_ATCC_199 EGALNGPSIMPGGPKKSYESLGPLLEEISAHVDGVPCCTHIGPDGSGHFV
Rv1844c|M.tuberculosis_H37Rv EGALNGPSIMPGGPAESYQSLGPLLEEISAHVDGVPCCTHIGPDGSGHFV
Mb1875c|M.bovis_AF2122/97 EGALNGPSIMPGGPAESYQSLGPLLEEISAHVDGVPCCTHIGPDGSGHFV
****:********. :** :***:**:********.*******.*:****
MMAR_2718|M.marinum_M KMVHNGIEYSDMQLIGEAYQLLRDGLGMSAPQIADVFSEWNKGDLDSYLV
MUL_3035|M.ulcerans_Agy99 KMVHNGIEYSDMQLIGEAYQLLRDGLGMSAPQIADVFSEWNKGDLDSYLV
MLBr_02065|M.leprae_Br4923 KMVHNGIEYSDMQLIGEAYQLLRDGLGMSAPQIADVFTEWNRGDLNSYLV
Mflv_3359|M.gilvum_PYR-GCK KMVHNGIEYSDMQLIGEAYQLLRDGLGKSAPEIADVFDEWNAGDLDSFLV
Mvan_3089|M.vanbaalenii_PYR-1 KMVHNGIEYSDMQLIGEAYQLLRDGLGKTAPEIADVFDEWNSGDLDSFLV
MAV_2871|M.avium_104 KMVHNGIEYSDMQLIGEAYQLLRDALGKTAEQIADVFDEWNSGDLDSFLV
TH_2383|M.thermoresistible__bu KMVHNGIEYSDMQLIGEAYQLLRDGLDLTAPQIADVFTEWNAGDLDSFLV
MSMEG_3632|M.smegmatis_MC2_155 KMVHNGIEYSDMQLIGEAYQLLRDGLGMSAGEIADVFEEWNRGDLDSFLV
MAB_2413c|M.abscessus_ATCC_199 KMVHNGIEYSDMQLIGEAYQLLRDGLGMSAPQIADVFTAWNKGELESYLI
Rv1844c|M.tuberculosis_H37Rv KMVHNGIEYSDMQLIGEAYQLMRDGLGLTAPAIADVFTEWNNGDLDSYLV
Mb1875c|M.bovis_AF2122/97 KMVHNGIEYSDMQLIGEAYQLMRDGLGLTAPAIADVFTEWNNGDLDSYLV
*********************:**.*. :* ***** ** *:*:*:*:
MMAR_2718|M.marinum_M EITAEVLRQTDAKTGKPLVDVILDQAEQKGTGRWTVKSALDLGVPVTGIA
MUL_3035|M.ulcerans_Agy99 EITAEVLRQTDAKTGKPLVDVILDQAEQKGTGRWTVKSALDLGVPVTGIA
MLBr_02065|M.leprae_Br4923 EITAEVLRQTDIKTGRPLVDVILDKAEQKGTGRWTVQSALDLGVPITGIA
Mflv_3359|M.gilvum_PYR-GCK EITAQVLRQTDATTGTPLVDVILDEAEQKGTGRWTVKSALDLGVPVTGIA
Mvan_3089|M.vanbaalenii_PYR-1 EITAQVLRQTDAKTGKPLVDLILDEAEQKGTGRWTVKSALDLGVPVTGIA
MAV_2871|M.avium_104 EITAQVLRQTDAKTGKPLVDLILDEAEQKGTGRWTVKSALDLGVPVTGIA
TH_2383|M.thermoresistible__bu EITAEVLRQIDAKTGKPLVDVIVDEAEQKGTGRWTVKSALDLGIPVTGIA
MSMEG_3632|M.smegmatis_MC2_155 EITAKVLRQTDAKTGKPLVDVIVDEAEQKGTGRWTVKSALDLGVPVTGIA
MAB_2413c|M.abscessus_ATCC_199 EITAEVLKQVDAKTGKPLVDVIVDAAEQKGTGRWTVKSALDLGVPVTGIA
Rv1844c|M.tuberculosis_H37Rv EITAEVLRQTDAKTGKPLVDVIVDRAEQKGTGRWTVKSALDLGVPVTGIA
Mb1875c|M.bovis_AF2122/97 EITAEVLRQTDAKTGKPLVDVIVDRAEQKGTGRWTVKSALDLGVPVTGIA
****:**:* * .** ****:*:* ***********:******:*:****
MMAR_2718|M.marinum_M EAVFARALSGSVQQRAAAVGLPSGRLGD-KPAD---PDTFTENVRRALYA
MUL_3035|M.ulcerans_Agy99 EAVFARALSGSVQQRAAAVGLPSGRLGD-KPAD---PDTFTENVRRALHA
MLBr_02065|M.leprae_Br4923 EAVFARALSGSVLQRKAAIGLASGKLGN-KPTD---RETFIEDVRQALYA
Mflv_3359|M.gilvum_PYR-GCK EAVFARALSGSVAQRKATTGLASGDLGE-KPGD---AEQFVEDIRQALYA
Mvan_3089|M.vanbaalenii_PYR-1 EAVFARALSGSVAQRRATTGLASGRLGERVPADGEAAAQFVEDIRQALYA
MAV_2871|M.avium_104 EAVFARALSGSVAQRRATTGLASGRLGE-KPSD---AAQFTEDIRQALYA
TH_2383|M.thermoresistible__bu EAVFARALSGSVAQRRATTGLASGTLSD-KPGD---AERFTEDVRRALYA
MSMEG_3632|M.smegmatis_MC2_155 EAVFARALSGSVPQRKATVGLASGALGA-TPSD---AADFIEAVRRALYA
MAB_2413c|M.abscessus_ATCC_199 EAVFARALSGSKPQRAASVGFASGQLGE-KPSD---AKQFVEDVRQALYA
Rv1844c|M.tuberculosis_H37Rv EAVFARALSGSVGQRSAASGLASGKLGE-QPAD---PATFTEDVRQALYA
Mb1875c|M.bovis_AF2122/97 EAVFARALSGSVGQRSAASGLASGKLGE-QPAD---PATFTEDVRQALYA
*********** ** *: *:.** *. * * * * :*:**:*
MMAR_2718|M.marinum_M SKIVAYAQGFNQIQAGSAEFGWDITPGDLATIWRGGCIIRAKFLNRIKEA
MUL_3035|M.ulcerans_Agy99 SKIVAYAQGFNQIQAGSAEFGWDITPDDLATIWRGGCIIRAKFLNRIKEA
MLBr_02065|M.leprae_Br4923 SKIVAYAQGFNHIQTGSTEFGWNITPGDLATIWRGGCIIRAKFLNRIKEA
Mflv_3359|M.gilvum_PYR-GCK SKIIAYAQGFNQIQAGSDEYDWDIKPGDMARIWRGGCIIRAKFLNRITDA
Mvan_3089|M.vanbaalenii_PYR-1 SKIIAYAQGFNQIQAGSAEYNWGITPGDMATIWRGGCIIRAKFLNRIKDA
MAV_2871|M.avium_104 SKIIAYAQGFNQIQAGSAEYGWDITPGDLATIWRGGCIIRAKFLNRIKDA
TH_2383|M.thermoresistible__bu SKIIAYAQGFNQIQAGSEEYNWNINPGDLATIWRGGCIIRAKFLNRIKEA
MSMEG_3632|M.smegmatis_MC2_155 SKIVAYAQGFNQIQAGSAEYNWNLTPGDLATIWRGGCIIRAKFLNRIKDA
MAB_2413c|M.abscessus_ATCC_199 SKIVAYAQGFNQIAAGSAEYGWNVSPGDLATIWRGGCIIRAQFLNRVKEA
Rv1844c|M.tuberculosis_H37Rv SKIVAYAQGFNQIQAGSAEFGWDITPGDLATIWRGGCIIRAKFLNHIKEA
Mb1875c|M.bovis_AF2122/97 SKIVAYAQGFNQIQAGSAEFGWDITPGDLATIWRGGCIIRAKFLNHIKEA
***:*******:* :** *:.*.:.*.*:* **********:***::.:*
MMAR_2718|M.marinum_M FDAEPALASLLVAPYFRSA------------------IESAVDSWR--RV
MUL_3035|M.ulcerans_Agy99 FDAEPALASLLVAPYFRSA------------------IESAVDSWR--RV
MLBr_02065|M.leprae_Br4923 FDAAPDLASLIVAPYFRSA------------------VESAIDSWR--RV
Mflv_3359|M.gilvum_PYR-GCK FDNDPGLPTLIVDPYFRDA------------------IEAAIDSWR--RV
Mvan_3089|M.vanbaalenii_PYR-1 FDNDPDLPTLIVDPYFRDA------------------IEAAVDSWR--RV
MAV_2871|M.avium_104 FDENPDLPTLIVAPYFRSA------------------IEAAIDGWR--RV
TH_2383|M.thermoresistible__bu FDADPDLPTLIVAPYFREA------------------IEAGVDSWR--RV
MSMEG_3632|M.smegmatis_MC2_155 YDAEPDLPTLLAAPYFRDA------------------VEAGIDSWR--RV
MAB_2413c|M.abscessus_ATCC_199 FADRPDLATLIAAPYFRDA------------------VEKGIDSWR--RV
Rv1844c|M.tuberculosis_H37Rv FDASPNLASLIVAPYFRGA------------------VESAIDSWR--RV
Mb1875c|M.bovis_AF2122/97 FDASPNLASLIVAPYFRAPSNRRSTVGGVWCRRRPNWVSRPRDSRRPCRI
: * *.:*:. **** . :. *. * *:
MMAR_2718|M.marinum_M VSTAAQLGIPTP--------------------------------------
MUL_3035|M.ulcerans_Agy99 VSTAAQLGIPTP--------------------------------------
MLBr_02065|M.leprae_Br4923 VSTATQLGIPNP--------------------------------------
Mflv_3359|M.gilvum_PYR-GCK VVKATELGIPIP--------------------------------------
Mvan_3089|M.vanbaalenii_PYR-1 VIKATELGIPIP--------------------------------------
MAV_2871|M.avium_104 VVTATRLGIPIP--------------------------------------
TH_2383|M.thermoresistible__bu VATATGLGVPVP--------------------------------------
MSMEG_3632|M.smegmatis_MC2_155 VVTATQLGIPVP--------------------------------------
MAB_2413c|M.abscessus_ATCC_199 VVTATQLGIPVP--------------------------------------
Rv1844c|M.tuberculosis_H37Rv VSTAAQLGIPTP--------------------------------------
Mb1875c|M.bovis_AF2122/97 TTRCAPRGCPLHSPRPSATSSAHTPTAGSTNQASSTHYGVQTAPKYRCSG
. .: * *
MMAR_2718|M.marinum_M -------GFSSALAYYDALRT---------------ERLPAALTQAQRDF
MUL_3035|M.ulcerans_Agy99 -------GFSSALAYYDALRT---------------ERLPAALTQAQRDF
MLBr_02065|M.leprae_Br4923 -------GFSSALSYYDALRT---------------ERLPAALTQAQRDF
Mflv_3359|M.gilvum_PYR-GCK -------GFSSALSYYDGLRT---------------ERLPAALTQGLRDF
Mvan_3089|M.vanbaalenii_PYR-1 -------GFSSALSYYDGLRT---------------ERLPAALTQGLRDF
MAV_2871|M.avium_104 -------GFSSALSYYDALRT---------------ERLPAALTQGLRDF
TH_2383|M.thermoresistible__bu -------GFASALSYYDALRT---------------ERLPAALIQGLRDY
MSMEG_3632|M.smegmatis_MC2_155 -------GFASALSYYDALRT---------------ERLPAALIQGLRDY
MAB_2413c|M.abscessus_ATCC_199 -------GFASSLSYYDGLRT---------------DRLPAALTQAQRDF
Rv1844c|M.tuberculosis_H37Rv -------GFSSALSYYDALRT---------------ARLPAALTQAQRDF
Mb1875c|M.bovis_AF2122/97 LELKGGKGVSDEISRRAPTRVRPDIQRRVHRSEPIRGRVALRRRFVHRRR
*.:. :: *. *:. *
MMAR_2718|M.marinum_M FGAHTYG-----------------------------------RID-----
MUL_3035|M.ulcerans_Agy99 FGAHTYG-----------------------------------RID-----
MLBr_02065|M.leprae_Br4923 FGAHSYG-----------------------------------RVD-----
Mflv_3359|M.gilvum_PYR-GCK FGAHTYG-----------------------------------RID-----
Mvan_3089|M.vanbaalenii_PYR-1 FGAHTYG-----------------------------------RID-----
MAV_2871|M.avium_104 FGAHTYG-----------------------------------RID-----
TH_2383|M.thermoresistible__bu FGAHTYG-----------------------------------RID-----
MSMEG_3632|M.smegmatis_MC2_155 FGAHTYG-----------------------------------RID-----
MAB_2413c|M.abscessus_ATCC_199 FGAHTYE-----------------------------------RID-----
Rv1844c|M.tuberculosis_H37Rv FGAHTYG-----------------------------------RID-----
Mb1875c|M.bovis_AF2122/97 LGHHHSGSGRQYDRGSRAADGRDGRPPRWHRNPAAGSADPGGKADGGVRQ
:* * : *
MMAR_2718|M.marinum_M ----------AAG--KFHTLWSADRSEVPA--------------------
MUL_3035|M.ulcerans_Agy99 ----------AAG--KFHTLWSADRSEVPA--------------------
MLBr_02065|M.leprae_Br4923 ----------ARG--KFHTLWSEDRSEVTV--------------------
Mflv_3359|M.gilvum_PYR-GCK ----------TDRERRFHTLWSGDRTEVEA--------------------
Mvan_3089|M.vanbaalenii_PYR-1 ----------TDPTRRFHTLWSGDRSEVEA--------------------
MAV_2871|M.avium_104 ----------EDPDKRFHTLWSADRREVPA--------------------
TH_2383|M.thermoresistible__bu ----------AEPGQKFHTLWSGDRSEVPA--------------------
MSMEG_3632|M.smegmatis_MC2_155 ----------AEPGKKFHTLWSGDHSEVEA--------------------
MAB_2413c|M.abscessus_ATCC_199 ----------APG--KFHTLWSGDRNEVEA--------------------
Rv1844c|M.tuberculosis_H37Rv ----------EPG--KFHTLWSSDRTEVPV--------------------
Mb1875c|M.bovis_AF2122/97 KPGPGARHPSDAGTRRFGVRRHGAHPQARTWRRGGHPRGSPDRIGARIVL
:* . : :. .
MMAR_2718|M.marinum_M --------------------------------------------------
MUL_3035|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02065|M.leprae_Br4923 --------------------------------------------------
Mflv_3359|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3089|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2871|M.avium_104 --------------------------------------------------
TH_2383|M.thermoresistible__bu --------------------------------------------------
MSMEG_3632|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2413c|M.abscessus_ATCC_199 --------------------------------------------------
Rv1844c|M.tuberculosis_H37Rv --------------------------------------------------
Mb1875c|M.bovis_AF2122/97 PGRGSLHPGARYRRDGLCDRSSGNRATQDLRPAGARPGRRCGADRRRRHV
MMAR_2718|M.marinum_M --------------------
MUL_3035|M.ulcerans_Agy99 --------------------
MLBr_02065|M.leprae_Br4923 --------------------
Mflv_3359|M.gilvum_PYR-GCK --------------------
Mvan_3089|M.vanbaalenii_PYR-1 --------------------
MAV_2871|M.avium_104 --------------------
TH_2383|M.thermoresistible__bu --------------------
MSMEG_3632|M.smegmatis_MC2_155 --------------------
MAB_2413c|M.abscessus_ATCC_199 --------------------
Rv1844c|M.tuberculosis_H37Rv --------------------
Mb1875c|M.bovis_AF2122/97 GGSAKPHRGYPRRYLHPGHR