For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQLGLVGLGKMGFNMRDRLRAGGHEVIGYDPRPEVTDVPSLKGLAEALDAPRVVWVMVPSGPITQQTIAE LSEELSEGDLVIDGGNSRFTEDKPNADLLAAKGIGYIDAGVSGGVWGKENGYGLMVGGSDEDVARAMPIF DTLRPEGERADGFVHAGPVGAGHYAKMIHNGIEYGLMHAYAEGYELLAAEPLITDVQAVLQAWTKGTVVR SWLLDLLAKALKEDPGFDAISGYTEDSGEGRWTVEEAINHRVPAPVIAASLFARFASRQEDSPAMKAVSA LRNQFGGHAVKRVGLSG
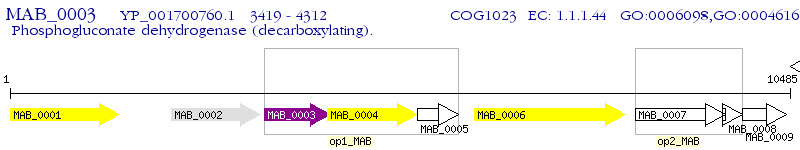
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0003 | gnD | - | 100% (297) | 6-phosphogluconate dehydrogenase-like protein |
| M. abscessus ATCC 19977 | MAB_2413c | - | 3e-39 | 33.68% (291) | 6-phosphogluconate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4486 | - | 1e-14 | 31.51% (219) | 3-hydroxyisobutyrate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1153 | gnd2 | 2e-77 | 46.29% (337) | 6-phosphogluconate dehydrogenase-like protein |
| M. gilvum PYR-GCK | Mflv_0825 | - | 1e-135 | 78.11% (297) | 6-phosphogluconate dehydrogenase-like protein |
| M. tuberculosis H37Rv | Rv1122 | gnd2 | 2e-77 | 46.29% (337) | 6-phosphogluconate dehydrogenase-like protein |
| M. leprae Br4923 | MLBr_02065 | gnd | 5e-37 | 32.30% (291) | 6-phosphogluconate dehydrogenase |
| M. marinum M | MMAR_4338 | gnd2 | 1e-75 | 45.72% (339) | 6-phosphogluconate dehydrogenase, decarboxylating Gnd2 |
| M. avium 104 | MAV_1253 | gnd | 1e-75 | 46.29% (337) | 6-phosphogluconate dehydrogenase-like protein |
| M. smegmatis MC2 155 | MSMEG_0002 | - | 1e-141 | 82.15% (297) | 6-phosphogluconate dehydrogenase-like protein |
| M. thermoresistible (build 8) | TH_3353 | - | 1e-132 | 77.44% (297) | PUTATIVE 6-phosphogluconate dehydrogenase (decarboxylating) |
| M. ulcerans Agy99 | MUL_0148 | gnd2 | 4e-75 | 45.43% (339) | 6-phosphogluconate dehydrogenase-like protein |
| M. vanbaalenii PYR-1 | Mvan_0003 | - | 1e-130 | 76.77% (297) | 6-phosphogluconate dehydrogenase-like protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1153|M.bovis_AF2122/97 ----------MQLGMIGLGRMGANIVRRLAKGGHDCVVYD----HDPDAV
Rv1122|M.tuberculosis_H37Rv ----------MQLGMIGLGRMGANIVRRLAKGGHDCVVYD----HDPDAV
MMAR_4338|M.marinum_M ----------MQLGMIGLGRMGANIVRRVVNGGHECVVYD----HNPDAV
MUL_0148|M.ulcerans_Agy99 ----------MQLGMIGLGRMGADIVRRVVNGGHECVVYD----HNPDAV
MAV_1253|M.avium_104 ----------MQLGMIGLGRMGANIVRRVVKGGHECVVYD----HNPDAV
MAB_0003|M.abscessus_ATCC_1997 ----------MQLGLVGLGKMGFNMRDRLRAGGHEVIGYD----PRPE--
MSMEG_0002|M.smegmatis_MC2_155 ----------MQLGLIGLGKMGFNMRERLREAGHEVIGYD----PRPE--
Mflv_0825|M.gilvum_PYR-GCK ----------MQLGLIGLGKMGFNMRARLRDGGHDVVGYD----PRPE--
Mvan_0003|M.vanbaalenii_PYR-1 ----------MQLGLIGLGKMGFNMRARLRDGGHDVVGFD----PRPE--
TH_3353|M.thermoresistible__bu ----------MQLGLVGLGKMGFNMRERLRAGGHEVIGYD----PRPE--
MLBr_02065|M.leprae_Br4923 MSAPESKTAIAQIGVTGLAVMGSNIARNFARHGYTVALHNRSIAKTDTLL
*:*: **. ** :: .. *: .:
Mb1153|M.bovis_AF2122/97 KAMAGEDRTTGVASLRELSQRLSAPRVVWVMVPAGNITTAVIEELANTLE
Rv1122|M.tuberculosis_H37Rv KAMAGEDRTTGVASLRELSQRLSAPRVVWVMVPAGNITTAVIEELANTLE
MMAR_4338|M.marinum_M KAMAGENNTTGVSSLSELRDKLSAPRVIWVMVPAGTITTGVIEELATTLD
MUL_0148|M.ulcerans_Agy99 KAMAGENNTTGVSSLSELRDKLSAPRVIWVMVPAGTITTGVIEELATTLD
MAV_1253|M.avium_104 KAMAGEEKTTAVSSLQELAAKLSAPRVVWVMVPAGTITTGVIEELATTLE
MAB_0003|M.abscessus_ATCC_1997 --------VTDVPSLKGLAEALDAPRVVWVMVPSGPITQQTIAELSEELS
MSMEG_0002|M.smegmatis_MC2_155 --------VSDVANLEELAGALSAPRVVWVMVPSGTITHSTIVELAGVLS
Mflv_0825|M.gilvum_PYR-GCK --------VSDVESLEELAQRLDAPRVVWVMVPSGTVTDSTITELAEVLS
Mvan_0003|M.vanbaalenii_PYR-1 --------VSDVASLADLAARLSAPRVVWVMVPSGTVTDSTISELAGVLS
TH_3353|M.thermoresistible__bu --------VTDVPSPAALTDALTAPRVVWVMVPSGSVTHDTIVALADVLS
MLBr_02065|M.leprae_Br4923 KEHGSEGNFVRTETIPEFLAALQTPRRVLIMVKAGDATDAVINELADVME
. . : * :** : :** :* * .* *: :.
Mb1153|M.bovis_AF2122/97 AGDIVIDGGNTYYRDDLRHEKLLFKKGIHLLDCGTSGGVWGRERGYCLMI
Rv1122|M.tuberculosis_H37Rv AGDIVIDGGNTYYRDDLRHEKLLFKKGIHLLDCGTSGGVWGRERGYCLMI
MMAR_4338|M.marinum_M AGDIVIDGGNTYYRDDIKHAKLLSGKGIHMLDCGTSGGVWGLDRGYCLMV
MUL_0148|M.ulcerans_Agy99 AGDIVIDGGNTYYRDDIKHAKLLSGKGIHMLDCGTSGGVWGLDRGYCLMV
MAV_1253|M.avium_104 PGDIVIDGGNSYYRDDIRHSKTLSEKGIRLLDCGTSGGVWGRERGYCLMI
MAB_0003|M.abscessus_ATCC_1997 EGDLVIDGGNSRFTEDKPNADLLAAKGIGYIDAGVSGGVWGKENGYGLMV
MSMEG_0002|M.smegmatis_MC2_155 PGDLVIDGGNSRYTEDGPHAELLGEKGISFIDAGVSGGIWGLTEGYGLMV
Mflv_0825|M.gilvum_PYR-GCK EGDLVIDGGNSRYTEDQPHAKMLAQKGIGFVDAGVSGGIWGLTEGYGLMV
Mvan_0003|M.vanbaalenii_PYR-1 PGDLVIDGGNSRYTEDGRHANLLAEKGIAFIDAGVSGGIWGLTEGYGLMV
TH_3353|M.thermoresistible__bu PGDLLIDGGNSRYTEDAKHQKLLGENGISFIDAGVSGGVWGLTDGYGLMV
MLBr_02065|M.leprae_Br4923 PSDIIIDGGNSLFTDTIRREKAMRERGLHFVGAGISGGEEGALNGPSIMP
.*::*****: : : . . : .*: :..* *** * * :*
Mb1153|M.bovis_AF2122/97 GGDGDAFARAEPIFATVAPGVAAAPRTPGRDGEVAPSEQGYLHCGPCGSG
Rv1122|M.tuberculosis_H37Rv GGDGDAFARAEPIFATVAPGVAAAPRTPGRDGEVAPSEQGYLHCGPCGSG
MMAR_4338|M.marinum_M GGEPDAFAHAEPILATVAPGVQAAPRTPGRDGEVAEAEKGYLHCGPTGAG
MUL_0148|M.ulcerans_Agy99 GGEPDAFAHAEPILATVAPGVQAAPRTPGRDGEVAEAEKGYLHCGPTGAG
MAV_1253|M.avium_104 GGDEEAFAHAEPIFATVAPGVDAAPRTPGRDGEVTRSEQGYLHCGPSGAG
MAB_0003|M.abscessus_ATCC_1997 GGSDEDVARAMPIFDTLRP--------EG------ERADGFVHAGPVGAG
MSMEG_0002|M.smegmatis_MC2_155 GGSDDDVARAMPIFDALRP--------PG------DVADGFVHAGPVGAG
Mflv_0825|M.gilvum_PYR-GCK GGSDADIARAMPIFDTLRP--------PG------PQEDGFVHVGPVGAG
Mvan_0003|M.vanbaalenii_PYR-1 GGSDADVARAMPIFETLRP--------PG------PLEDGFVHVGPVGAG
TH_3353|M.thermoresistible__bu GGSADDVARAMPIFDALRP--------PG------PRADGFVHAGPTGAG
MLBr_02065|M.leprae_Br4923 GGPAESYTSLGPLLEEISAH--------------VDGVSCCTHIGPGGSG
** : *:: : . . * ** *:*
Mb1153|M.bovis_AF2122/97 HFVKMVHNGIEYGMMASLAEGLNILRNADVGTRVQHGDAETAPLPNPECY
Rv1122|M.tuberculosis_H37Rv HFVKMVHNGIEYGMMASLAEGLNILRNADVGTRVQHGDAETAPLPNPECY
MMAR_4338|M.marinum_M HFVKMVHNGIEYGMMASLAEGLNILRNADVGTRVQQGDAETAPLANPEYY
MUL_0148|M.ulcerans_Agy99 HFVKMVHNGIEYGMMASLAEGLNILRNADVGTRVQQGDAETAPLANPEYY
MAV_1253|M.avium_104 HFVKMVHNGIEYGMMASLAEGLNVLRNADIGKHVQEGDAETAPLSNPEFY
MAB_0003|M.abscessus_ATCC_1997 HYAKMIHNGIEYGLMHAYAEGYELLAAEPLITDVQ---------------
MSMEG_0002|M.smegmatis_MC2_155 HYAKMVHNGIEYGLMHAYAEGYELLAAEELIKDPQ---------------
Mflv_0825|M.gilvum_PYR-GCK HFAKMVHNGIEYALMTAYAEGYELLAAEELVKDPQ---------------
Mvan_0003|M.vanbaalenii_PYR-1 HFAKMVHNGIEYALMTAYAEGYEMLAAEDLVKDPQ---------------
TH_3353|M.thermoresistible__bu HFAKMVHNGVEYALMTAYAEGYELLSAADLITDPQ---------------
MLBr_02065|M.leprae_Br4923 HFVKMVHNGIEYSDMQLIGEAYQLLRDGLGMSAPQ---------------
*:.**:***:**. * .*. ::* . *
Mb1153|M.bovis_AF2122/97 QYDFDIPEVAEVWRRGSVIGSWLLDLTAIALRES------PDLAEFSGRV
Rv1122|M.tuberculosis_H37Rv QYDFDIPEVAEVWRRGSVIGSWLLDLTAIALRES------PDLAEFSGRV
MMAR_4338|M.marinum_M QYDIDIPEVAEVWRRGSVIGSWLLDLTAIALHES------PELQEFSGRV
MUL_0148|M.ulcerans_Agy99 QYDIDIPEVAEVWRRGSVIGSWLLDLTAIALHES------PELQEFSGRV
MAV_1253|M.avium_104 QYDFDIPEVAEVWRRGSVIGSWLLDLTAIALHES------PELKEFSGRV
MAB_0003|M.abscessus_ATCC_1997 -------AVLQAWTKGTVVRSWLLDLLAKALKED------PGFDAISGYT
MSMEG_0002|M.smegmatis_MC2_155 -------AVIQAWSNGTVVRSWLQSLLAKALKED------PGFAEISGYT
Mflv_0825|M.gilvum_PYR-GCK -------AVYQAWTNGTVVRSWLQQLLAKALKED------PDLSEISGYT
Mvan_0003|M.vanbaalenii_PYR-1 -------AVYQAWTNGTVVRSWLQQLLAKALKED------PSLSDISGYT
TH_3353|M.thermoresistible__bu -------AVYQAWTNGTVVRSWLQQLLAKALEED------PGFSRITGYT
MLBr_02065|M.leprae_Br4923 -----IADVFTEWNRGDLN-SYLVEITAEVLRQTDIKTGRPLVDVILDKA
* * .* : *:* .: * .*.: * . : . .
Mb1153|M.bovis_AF2122/97 SDSGEGRWTAIAAIDEGVPAPVLTTALQSRFAS-----------------
Rv1122|M.tuberculosis_H37Rv SDSGEGRWTAIAAIDEGVPAPVLTTALQSRFAS-----------------
MMAR_4338|M.marinum_M SDSGEGRWTSIAAIDEGVPTPVLTTALQSRFAS-----------------
MUL_0148|M.ulcerans_Agy99 SDSGEGRWTSIAAIDEGVPTPVLTTALQSRFAS-----------------
MAV_1253|M.avium_104 SDSGEGRWTSIAAIDEGVPTPVLTTALQSRFAS-----------------
MAB_0003|M.abscessus_ATCC_1997 EDSGEGRWTVEEAINHRVPAPVIAASLFARFAS-----------------
MSMEG_0002|M.smegmatis_MC2_155 EDSGEGRWTVEEAIAHRVPMPVIAASLFARFAS-----------------
Mflv_0825|M.gilvum_PYR-GCK EDSGEGRWTVEEAIRLRVPVPGIAASLFARFLS-----------------
Mvan_0003|M.vanbaalenii_PYR-1 EDSGEGRWTVEEAIRLRVPVPGIAASLFARFLS-----------------
TH_3353|M.thermoresistible__bu EDSGEGRWTVEEAIRLRVPVPSIAASLFARFLS-----------------
MLBr_02065|M.leprae_Br4923 EQKGTGRWTVQSALDLGVPITGIAEAVFARALSGSVLQRKAAIGLASGKL
.:.* **** *: ** . :: :: :* *
Mb1153|M.bovis_AF2122/97 --------------------------------------------------
Rv1122|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4338|M.marinum_M --------------------------------------------------
MUL_0148|M.ulcerans_Agy99 --------------------------------------------------
MAV_1253|M.avium_104 --------------------------------------------------
MAB_0003|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_0002|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0825|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0003|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3353|M.thermoresistible__bu --------------------------------------------------
MLBr_02065|M.leprae_Br4923 GNKPTDRETFIEDVRQALYASKIVAYAQGFNHIQTGSTEFGWNITPGDLA
Mb1153|M.bovis_AF2122/97 --------------------------------------------------
Rv1122|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4338|M.marinum_M --------------------------------------------------
MUL_0148|M.ulcerans_Agy99 --------------------------------------------------
MAV_1253|M.avium_104 --------------------------------------------------
MAB_0003|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_0002|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0825|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0003|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3353|M.thermoresistible__bu --------------------------------------------------
MLBr_02065|M.leprae_Br4923 TIWRGGCIIRAKFLNRIKEAFDAAPDLASLIVAPYFRSAVESAIDSWRRV
Mb1153|M.bovis_AF2122/97 -----------------------RDLDDFANKALSAMRKQFGGHAEKPAN
Rv1122|M.tuberculosis_H37Rv -----------------------RDLDDFANKALSAMRKQFGGHAEKPAN
MMAR_4338|M.marinum_M -----------------------RDLDDFANKALSAMRKQFGGHAEKPVN
MUL_0148|M.ulcerans_Agy99 -----------------------RDLDDFANKALSAMRKQFGGHAEKPVN
MAV_1253|M.avium_104 -----------------------RHLDDFANKTLSAMRKQFGGHAEKPAN
MAB_0003|M.abscessus_ATCC_1997 -----------------------RQEDSPAMKAVSALRNQFGGHAVKRVG
MSMEG_0002|M.smegmatis_MC2_155 -----------------------RQEDSPTMKAVSALRNQFGGHAVKRIS
Mflv_0825|M.gilvum_PYR-GCK -----------------------RQDESPTMRAVAALRNQFGGHAVKRVS
Mvan_0003|M.vanbaalenii_PYR-1 -----------------------RQDESPTMKAVAALRNQFGGHAVKRVS
TH_3353|M.thermoresistible__bu -----------------------RQDESPTMKAVAALRQQFGGHAVKRVS
MLBr_02065|M.leprae_Br4923 VSTATQLGIPNPGFSSALSYYDALRTERLPAALTQAQRDFFGAHSYGRVD
: . * *. **.*: .
Mb1153|M.bovis_AF2122/97 ------------------
Rv1122|M.tuberculosis_H37Rv ------------------
MMAR_4338|M.marinum_M ------------------
MUL_0148|M.ulcerans_Agy99 ------------------
MAV_1253|M.avium_104 ------------------
MAB_0003|M.abscessus_ATCC_1997 LSG---------------
MSMEG_0002|M.smegmatis_MC2_155 ESG---------------
Mflv_0825|M.gilvum_PYR-GCK ESG---------------
Mvan_0003|M.vanbaalenii_PYR-1 ESG---------------
TH_3353|M.thermoresistible__bu ESG---------------
MLBr_02065|M.leprae_Br4923 ARGKFHTLWSEDRSEVTV