For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSMRRHIIVSGDDALANTIVEELNSAGVTILKLASDELNDAQLAQAIAVICAGDDDARNLEIALLARRAD PHVRVVARLANDVLREAMAADNGPGAILDVADLAAPSVVEACLARTVHPFEAAGIEFVVWRAEAPRDATL REIYGDLAPVAVVRGKNSPTPGEVLTCPGRDLRVHAGDETAMVGTAEDLAARGIKVPRPTSTRFRLLRLR QISDAARALRNDVNPMFFKALAAMATLLAVSTTLLRFAYRHPPGMTWLDALYFSTETIATVGYGDFSFSH QHPWLRVFSITLMFAGVTTTAILVAFIADLLLSRRFTRSAGRRRVRQLRNHVIVVGLGSFGIRVVRDLIA AGYDVAVIELDENSRFLESAAELDVPVIFGDATLPQTLESACIAHARAMAVLTQDDMVNIETGIVLREML GAVKANRRDIPLVLRVYDRELGGAVAQRFDFESVRSTVELAAPWFIGAAMGLQVLGTFSVGQSSFMVGGM HVSAQSELDGLSMRELVTGTRVIAITRPDEPPQLHPRRDTRLRAGDTAYLVGPYRELLATLRKGQPPQQP ASGDGHWPEVATVDNVGPLDRPN
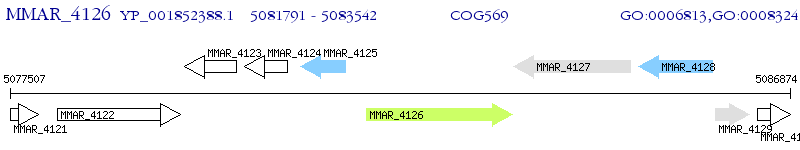
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4126 | - | - | 100% (583) | putative transport protein |
| M. marinum M | MMAR_1360 | - | 4e-09 | 26.67% (195) | transmembrane cation transporter |
| M. marinum M | MMAR_2023 | ceoB | 1e-04 | 26.29% (213) | TRK system potassium uptake protein CeoB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3225c | - | 3e-11 | 27.72% (202) | transmembrane cation transporter |
| M. gilvum PYR-GCK | Mflv_4663 | - | 4e-11 | 29.44% (197) | TrkA domain-containing protein |
| M. tuberculosis H37Rv | Rv3200c | - | 3e-11 | 27.72% (202) | transmembrane cation transporter |
| M. leprae Br4923 | MLBr_01763 | - | 5e-13 | 54.29% (70) | hypothetical protein MLBr_01763 |
| M. abscessus ATCC 19977 | MAB_3514c | - | 5e-11 | 25.15% (167) | transmembrane cation transporter |
| M. avium 104 | MAV_1443 | - | 0.0 | 73.27% (565) | TrkA domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_1945 | - | 3e-14 | 29.31% (174) | ion channel membrane protein |
| M. thermoresistible (build 8) | TH_2097 | - | 6e-14 | 28.08% (203) | POSSIBLE TRANSMEMBRANE CATION TRANSPORTER |
| M. ulcerans Agy99 | MUL_2519 | - | 4e-10 | 27.18% (195) | transmembrane cation transporter |
| M. vanbaalenii PYR-1 | Mvan_1804 | - | 2e-11 | 28.57% (175) | TrkA domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1945|M.smegmatis_MC2_155 --------------------------------------------------
TH_2097|M.thermoresistible__bu --------------------------------------------------
Mflv_4663|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1804|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3514c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3225c|M.bovis_AF2122/97 --------------------------------------------------
Rv3200c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2519|M.ulcerans_Agy99 --------------------------------------------------
MMAR_4126|M.marinum_M -------MSMRRHIIVSGDDALANTIVEELNSAGVTILKLASDELNDAQL
MAV_1443|M.avium_104 MRDDSPRMSPRRHIIVSGDDVLATTIAEELNRAGATIVKLPSEELAGADL
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MSMEG_1945|M.smegmatis_MC2_155 ------------------------------MAKGRLRRRLAAIDSNLTSQ
TH_2097|M.thermoresistible__bu -----------------------------------LRRRLAAIEQDLTSQ
Mflv_4663|M.gilvum_PYR-GCK ------------------------------MAEGRLRRRLRRFDQSLADM
Mvan_1804|M.vanbaalenii_PYR-1 ------------------------------MAEGRLRRRLSRFDQSLAEM
MAB_3514c|M.abscessus_ATCC_199 -------------------------------MAGKLRQRFRGIDQALTAQ
Mb3225c|M.bovis_AF2122/97 -------------------------------MAG-SWRRLRGLDEKLTAQ
Rv3200c|M.tuberculosis_H37Rv -------------------------------MAG-SWRRLRGLNEKLTAQ
MUL_2519|M.ulcerans_Agy99 ------------------------------MAKG-SWRRLRRLDERLTAQ
MMAR_4126|M.marinum_M AQAIAVICAGDDDARNLEIALLARRADPHVRVVARLANDVLREAMAADNG
MAV_1443|M.avium_104 ARASAIVCAGRDDAKNLEIALLARKTNPHVRVVARLGNDVLRGAVAADNG
MLBr_01763|M.leprae_Br4923 -------------------------------MVARLASEVLRETVADDDR
.
MSMEG_1945|M.smegmatis_MC2_155 PDAALVDVLRIPEPFISPSRKIAMRLLYATGALFAAVLIVYLDRHGYRDI
TH_2097|M.thermoresistible__bu PEARLVDILRIPEPFVSPTQRIVRRIIYATLALMAAVVIVYLDRDGYRDV
Mflv_4663|M.gilvum_PYR-GCK PVHALTDRVQIPTETVSPARRITVRVIYALGALFAAVLIVYLDRDGYRDS
Mvan_1804|M.vanbaalenii_PYR-1 PVHSLTDRVQIPTETVSPARRITVRVIYALAALFAAVLIVYMDRDGYRDS
MAB_3514c|M.abscessus_ATCC_199 PDHALVGVLRIPENAASPWRAIGKRILIAVGTLFAAVLVVYADRSGYRDV
Mb3225c|M.bovis_AF2122/97 PGYALVGVLRIPQRRASPARVISRRVVVAVVALLLTAGIVYVDRDGYLDA
Rv3200c|M.tuberculosis_H37Rv PGYALVGVLRIPQRRASPARVISRRVVVAVVALLLTAGIVYVDRDGYLDA
MUL_2519|M.ulcerans_Agy99 PSYALVGVLRIPQGQASPARIITRRVTIALAALLAGAITVYLDRDGYHDS
MMAR_4126|M.marinum_M PGAILDVADLAAPSVVEACLARTVHPFEAAGIEFVVWRAEAPRDATLREI
MAV_1443|M.avium_104 PGAILDVADLAAPSVVEACLSSSTHPVRAAGIDFLVSGAEAPRDATLREI
MLBr_01763|M.leprae_Br4923 SSAIFDVAAPATSVFVETFLGRTTHPVGAAGIEFMALAPKQSRDVTLREI
. : . .. : * : :
MSMEG_1945|M.smegmatis_MC2_155 NATPD---------------------------------------------
TH_2097|M.thermoresistible__bu QGNE----------------------------------------------
Mflv_4663|M.gilvum_PYR-GCK QENP----------------------------------------------
Mvan_1804|M.vanbaalenii_PYR-1 QENQ----------------------------------------------
MAB_3514c|M.abscessus_ATCC_199 ASTPE---------------------------------------------
Mb3225c|M.bovis_AF2122/97 QGDR----------------------------------------------
Rv3200c|M.tuberculosis_H37Rv QGDR----------------------------------------------
MUL_2519|M.ulcerans_Agy99 QGDE----------------------------------------------
MMAR_4126|M.marinum_M YGDLAPVAVVRGKNSPTPGEVLTCPGRDLRVHAGDETAMVGTAEDLAARG
MAV_1443|M.avium_104 YGDLAPVAVIHGNDGATPGEVVPCPGRDHRVRAGDWTAMIGSADELAARG
MLBr_01763|M.leprae_Br4923 YH------------------------------------------------
MSMEG_1945|M.smegmatis_MC2_155 --------------------------------------------------
TH_2097|M.thermoresistible__bu --------------------------------------------------
Mflv_4663|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1804|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3514c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3225c|M.bovis_AF2122/97 --------------------------------------------------
Rv3200c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2519|M.ulcerans_Agy99 --------------------------------------------------
MMAR_4126|M.marinum_M IKVPRPTSTRFRLLRLRQISDAARALRNDVNPMFFKALAAMATLLAVSTT
MAV_1443|M.avium_104 IRTPRPPATRSRQTWLRRVLDAARAMRDDVNPMLFPAILLALTLLLVSTV
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MSMEG_1945|M.smegmatis_MC2_155 -------ENDPLSFLDCLYYATVSLSTTGYGDITPYTESARLVNVLVITP
TH_2097|M.thermoresistible__bu -----------LSFLDCLYYATVSLSTTGYGDITPVTPEARLINVLVITP
Mflv_4663|M.gilvum_PYR-GCK -----------LSFLDCLYYATVSLSTTGYGDITPITPEARLINVLVITP
Mvan_1804|M.vanbaalenii_PYR-1 -----------LSFLDCLYYATVSLSTTGYGDITPITPTARLVNVLVITP
MAB_3514c|M.abscessus_ATCC_199 -------ESDPLSFLDCFYYATVSLSTTGYGDITPYTEGARLVNILVITP
Mb3225c|M.bovis_AF2122/97 -----------LTFLDCLYYAAVTLSTTGYGDITPISEFARAINIFVITP
Rv3200c|M.tuberculosis_H37Rv -----------LTFLDCLYYAAVTLSTTGYGDITPISEFARAINIFVITP
MUL_2519|M.ulcerans_Agy99 -----------LSFLDCLYYSAVTLSTTGYGDITPISELARAANVLIITP
MMAR_4126|M.marinum_M LLRFAYRHPPGMTWLDALYFSTETIATVGYGDFSFSHQHPWLRVFSITLM
MAV_1443|M.avium_104 IVHFSYTKPR-LSWLDALYFTAETITTVGYGEFTFAHQSAWLRIFAVGLM
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MSMEG_1945|M.smegmatis_MC2_155 LR-VAFLIVLIGTTVETLTTQSRQALKIQRWRSKVRNHTVVIGYGTKGRT
TH_2097|M.thermoresistible__bu LR-VAFLIVLIGTTVETLTSQSRAALKIQRWRSRVRNHTVVIGYGTKGKT
Mflv_4663|M.gilvum_PYR-GCK LR-VAFLIVLIGTTVETLTTQSRQVYQIQRWRSRLRNHIIIVGYGTKGRT
Mvan_1804|M.vanbaalenii_PYR-1 LR-VAFLIVLIGTTVETLTTQSRQVYQIQRWRSRLRNHIIIVGYGTKGRT
MAB_3514c|M.abscessus_ATCC_199 LR-LLFLIVLVGTTVEALTERSRQALKIQRWRARVRNHTVVVGYGTKGKT
Mb3225c|M.bovis_AF2122/97 LR-IAFLILLVGTTLEVLTETSRQAYKIQRWRSRVRNHTVVIGYGTKGKT
Rv3200c|M.tuberculosis_H37Rv LR-IAFLILLVGTTLEVLTETSRQAYKIQRWRSRVRNHTVVIGYGTKGKT
MUL_2519|M.ulcerans_Agy99 LR-IAFLILLVGTTLEVVTETSRQALKIRRWRSRVRNHTVVIGYGTKGKT
MMAR_4126|M.marinum_M FAGVTTTAILVAFIADLLLSRRFTRSAGRRRVRQLRNHVIVVGLGSFGIR
MAV_1443|M.avium_104 FAGVTTTALLVAFLADLLLSRRFLQSAGLRRARHLRNHIIVVGLGSFGSR
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MSMEG_1945|M.smegmatis_MC2_155 AVAAMVGDDVAPADIVVVDENASALERARSAGLVTVHGDATKSDILRLAS
TH_2097|M.thermoresistible__bu AVAAMVEDEVAPADIVVVDDDPTALERARAAGLVTVHGDATKAEVLRLAS
Mflv_4663|M.gilvum_PYR-GCK AAAAMVGDEIAPGDIVVVDEDTSALDRAKSAGLVTVRGTATDSEVLRLAG
Mvan_1804|M.vanbaalenii_PYR-1 AAAAMVGDEVAPADIVVVDEDAAALERAKSAGLVTVRGSATDSEVLRLAG
MAB_3514c|M.abscessus_ATCC_199 AVQAMLSDGAAPSEIVVVDEDQMALDAAAAADLVTVRGSATKSDVLRLAG
Mb3225c|M.bovis_AF2122/97 AVAAMVSDELVPGEIVVVDTDSGVLERAAAAGLVTVHGDATKSDVLRLAG
Rv3200c|M.tuberculosis_H37Rv AVAAMVSDELVPGEIVVVDTDSGVLERAAAAGLVTVHGDATKSDVLRLAG
MUL_2519|M.ulcerans_Agy99 AISAMLGDEAVPGEIVVVDTDRGALDRAAAAGLVIVHGDATKSDVLRLAG
MMAR_4126|M.marinum_M VVRDLIAAGYD-VAVIELDENSRFLESAAELDVPVIFGDATLPQTLESAC
MAV_1443|M.avium_104 VVADLTAAGYD-VAVIERDENNRFLSTAAELDVPVIFGDATLRQTLESAR
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MSMEG_1945|M.smegmatis_MC2_155 AQHAKAIIVATDDDASAVLVTLTARELAPKAKIIAAAREAENQHLLRQSG
TH_2097|M.thermoresistible__bu VQHAKSIIVATDNDASAVLVTLTARELAPNAKIIAAARESDNQHLLKQSG
Mflv_4663|M.gilvum_PYR-GCK AQHAKTIIVAANRDDTAVLVTLTARELAPRAQIIAAVRETDNEHLLKQSG
Mvan_1804|M.vanbaalenii_PYR-1 AQHAKSIIVATNRDDTAVLVTLTARELAPNAKIIAAVREAENQHLLEQSG
MAB_3514c|M.abscessus_ATCC_199 VQNAKSIIVAANRDDTSVLVTLTARELAPNAKIVAAIREAENVHLLRQSG
Mb3225c|M.bovis_AF2122/97 TQHASSIIVATSRDDTAVLVTLTAREIAPKAKIVASIREAENQHLLRQSG
Rv3200c|M.tuberculosis_H37Rv TQHASSIIVATSRDDTAVLVTLTAREIAPKAKIVASIREAENQHLLRQSG
MUL_2519|M.ulcerans_Agy99 AQHAASIIVAASRDDTAALVTLTAREISPKSQIVASIREAENQHLLEQSG
MMAR_4126|M.marinum_M IAHARAMAVLTQDDMVNIETGIVLREMLG---AVKANRRDIPLVLRVYDR
MAV_1443|M.avium_104 VDRARAVAVLTQDDMVNIEIGIVLREMLGPRVMPEVNRPDVPIVLRIYDR
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MSMEG_1945|M.smegmatis_MC2_155 ADSTVVSS------------ETAGRLLGIATQTPSVVEMMEDLLTPDAGF
TH_2097|M.thermoresistible__bu ADSTVVSS------------ETAGRLLGVATQTPSVVAIIEDLLTPDAGF
Mflv_4663|M.gilvum_PYR-GCK ADSTVVTS------------ETAGRLLGIAVQTPSVVEMMEDLLTPDRGF
Mvan_1804|M.vanbaalenii_PYR-1 ADSTVVTS------------ETAGRLLGIAVQTPSVVEMMEDLLTPDRGF
MAB_3514c|M.abscessus_ATCC_199 ADSVVVSS------------ETAGRLLGIATTTPRVVEMIEDLLTPEAGF
Mb3225c|M.bovis_AF2122/97 ADTVVVSS------------ETAGRLLGIATTTPSVVEMIEDLLTPEAGL
Rv3200c|M.tuberculosis_H37Rv ADTVVVSS------------ETAGRLLGIATTTPSVVEMIEDLLTPEAGL
MUL_2519|M.ulcerans_Agy99 ADSVVVSS------------ETAGRLLGLATTTPSVVQMIEDLLTPDAGL
MMAR_4126|M.marinum_M ELGGAVAQRFDFESVRSTVELAAPWFIGAAMGLQVLGTFSVGQSSFMVGG
MAV_1443|M.avium_104 TLGDAVAKRFGFENVRSTVDLAAPWFIGAAMGLQVLGTFSVGQRSFMVGA
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MSMEG_1945|M.smegmatis_MC2_155 AIAEREVSPKEEGGSPRHLHDIVLGVVRGGRLVRVDDPEVDALEAGDRLL
TH_2097|M.thermoresistible__bu SIAEREVTPKEVGGSPRHLQDIVLGVVRDGHLYRVDEPEVDAVEAGDRLL
Mflv_4663|M.gilvum_PYR-GCK AIAEREVSPKEAGGSPRHQTDIVLGVVRNKKLIRVDDPEVDALELGDRLL
Mvan_1804|M.vanbaalenii_PYR-1 AIAEREVSPKEAGSSPRHQTDIVLGVVRDNKLIRVDEPEVDALELGDRLL
MAB_3514c|M.abscessus_ATCC_199 AIAEREVERSEVGGSPRHLSDIVLGVVRDGILHRVDAPEVDSIEATDRLL
Mb3225c|M.bovis_AF2122/97 AVAEREVEQAEVGGSPRHLRDIVLGVVRDGQLLRIGAPEVDAIEASDRLL
Rv3200c|M.tuberculosis_H37Rv AVAEREVEQAEVGGSPRHLRDIVLGVVRDGQLLRIGAPEVDAIEASDRLL
MUL_2519|M.ulcerans_Agy99 AIAEREVEQSEIGGSPRHLRDIVLGVVRGGQLLRIGAPEVDAIEANDRLL
MMAR_4126|M.marinum_M MHVSAQSELDGLSMRELVTGTRVIAITRPDEPPQLHPRRDTRLRAGDTAY
MAV_1443|M.avium_104 MHVAAGSELDGLRMFEMSTQTRVIAITRRDTPVELHPRRDAWLRAGDTVY
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MSMEG_1945|M.smegmatis_MC2_155 YIRSADAER-----------------------------------
TH_2097|M.thermoresistible__bu YIRISDGDR-----------------------------------
Mflv_4663|M.gilvum_PYR-GCK YIRSADEDR-----------------------------------
Mvan_1804|M.vanbaalenii_PYR-1 YIRSADEER-----------------------------------
MAB_3514c|M.abscessus_ATCC_199 YVRNAGA-------------------------------------
Mb3225c|M.bovis_AF2122/97 YIRQVGR-------------------------------------
Rv3200c|M.tuberculosis_H37Rv YIRQVGR-------------------------------------
MUL_2519|M.ulcerans_Agy99 YIRNTGALVAIASAAQPGAAGRHQGGGVAIVSSAQPGCQGGES-
MMAR_4126|M.marinum_M LVGPYRELLATLRKGQPPQQPASGDGHWPEVATVDNVGPLDRPN
MAV_1443|M.avium_104 LVGPYRELLETLRKGQPPQQPANEERPADKATT-----------
MLBr_01763|M.leprae_Br4923 --------------------------------------------