For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVARLASEVLRETVADDDRSSAIFDVAAPATSVFVETFLGRTTHPVGAAGIEFMALAPKQSRDVTLREIY H
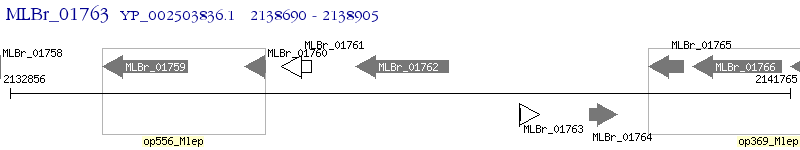
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01763 | - | - | 100% (71) | hypothetical protein MLBr_01763 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4126 | - | 1e-13 | 54.29% (70) | putative transport protein |
| M. avium 104 | MAV_1443 | - | 1e-12 | 51.43% (70) | TrkA domain-containing protein |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4126|M.marinum_M -------MSMRRHIIVSGDDALANTIVEELNSAGVTILKLASDELNDAQL
MAV_1443|M.avium_104 MRDDSPRMSPRRHIIVSGDDVLATTIAEELNRAGATIVKLPSEELAGADL
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MMAR_4126|M.marinum_M AQAIAVICAGDDDARNLEIALLARRADPHVRVVARLANDVLREAMAADNG
MAV_1443|M.avium_104 ARASAIVCAGRDDAKNLEIALLARKTNPHVRVVARLGNDVLRGAVAADNG
MLBr_01763|M.leprae_Br4923 -------------------------------MVARLASEVLRETVADDDR
:****..:*** ::* *:
MMAR_4126|M.marinum_M PGAILDVADLAAPSVVEACLARTVHPFEAAGIEFVVWRAEAPRDATLREI
MAV_1443|M.avium_104 PGAILDVADLAAPSVVEACLSSSTHPVRAAGIDFLVSGAEAPRDATLREI
MLBr_01763|M.leprae_Br4923 SSAIFDVAAPATSVFVETFLGRTTHPVGAAGIEFMALAPKQSRDVTLREI
..**:*** *:. .**: *. :.**. ****:*:. .: .**.*****
MMAR_4126|M.marinum_M YGDLAPVAVVRGKNSPTPGEVLTCPGRDLRVHAGDETAMVGTAEDLAARG
MAV_1443|M.avium_104 YGDLAPVAVIHGNDGATPGEVVPCPGRDHRVRAGDWTAMIGSADELAARG
MLBr_01763|M.leprae_Br4923 YH------------------------------------------------
*
MMAR_4126|M.marinum_M IKVPRPTSTRFRLLRLRQISDAARALRNDVNPMFFKALAAMATLLAVSTT
MAV_1443|M.avium_104 IRTPRPPATRSRQTWLRRVLDAARAMRDDVNPMLFPAILLALTLLLVSTV
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MMAR_4126|M.marinum_M LLRFAYRHPPGMTWLDALYFSTETIATVGYGDFSFSHQHPWLRVFSITLM
MAV_1443|M.avium_104 IVHFSYTKPR-LSWLDALYFTAETITTVGYGEFTFAHQSAWLRIFAVGLM
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MMAR_4126|M.marinum_M FAGVTTTAILVAFIADLLLSRRFTRSAGRRRVRQLRNHVIVVGLGSFGIR
MAV_1443|M.avium_104 FAGVTTTALLVAFLADLLLSRRFLQSAGLRRARHLRNHIIVVGLGSFGSR
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MMAR_4126|M.marinum_M VVRDLIAAGYDVAVIELDENSRFLESAAELDVPVIFGDATLPQTLESACI
MAV_1443|M.avium_104 VVADLTAAGYDVAVIERDENNRFLSTAAELDVPVIFGDATLRQTLESARV
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MMAR_4126|M.marinum_M AHARAMAVLTQDDMVNIETGIVLREMLG---AVKANRRDIPLVLRVYDRE
MAV_1443|M.avium_104 DRARAVAVLTQDDMVNIEIGIVLREMLGPRVMPEVNRPDVPIVLRIYDRT
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MMAR_4126|M.marinum_M LGGAVAQRFDFESVRSTVELAAPWFIGAAMGLQVLGTFSVGQSSFMVGGM
MAV_1443|M.avium_104 LGDAVAKRFGFENVRSTVDLAAPWFIGAAMGLQVLGTFSVGQRSFMVGAM
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MMAR_4126|M.marinum_M HVSAQSELDGLSMRELVTGTRVIAITRPDEPPQLHPRRDTRLRAGDTAYL
MAV_1443|M.avium_104 HVAAGSELDGLRMFEMSTQTRVIAITRRDTPVELHPRRDAWLRAGDTVYL
MLBr_01763|M.leprae_Br4923 --------------------------------------------------
MMAR_4126|M.marinum_M VGPYRELLATLRKGQPPQQPASGDGHWPEVATVDNVGPLDRPN
MAV_1443|M.avium_104 VGPYRELLETLRKGQPPQQPANEERPADKATT-----------
MLBr_01763|M.leprae_Br4923 -------------------------------------------