For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGKLRQRFRGIDQALTAQPDHALVGVLRIPENAASPWRAIGKRILIAVGTLFAAVLVVYADRSGYRDVAS TPEESDPLSFLDCFYYATVSLSTTGYGDITPYTEGARLVNILVITPLRLLFLIVLVGTTVEALTERSRQA LKIQRWRARVRNHTVVVGYGTKGKTAVQAMLSDGAAPSEIVVVDEDQMALDAAAAADLVTVRGSATKSDV LRLAGVQNAKSIIVAANRDDTSVLVTLTARELAPNAKIVAAIREAENVHLLRQSGADSVVVSSETAGRLL GIATTTPRVVEMIEDLLTPEAGFAIAEREVERSEVGGSPRHLSDIVLGVVRDGILHRVDAPEVDSIEATD RLLYVRNAGA*
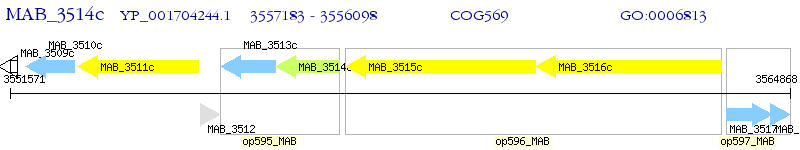
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3514c | - | - | 100% (361) | transmembrane cation transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3225c | - | 1e-140 | 72.22% (360) | transmembrane cation transporter |
| M. gilvum PYR-GCK | Mflv_4663 | - | 1e-133 | 67.79% (357) | TrkA domain-containing protein |
| M. tuberculosis H37Rv | Rv3200c | - | 1e-140 | 71.94% (360) | transmembrane cation transporter |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1360 | - | 1e-139 | 71.47% (354) | transmembrane cation transporter |
| M. avium 104 | MAV_4146 | - | 1e-136 | 70.06% (354) | ion channel membrane protein |
| M. smegmatis MC2 155 | MSMEG_1945 | - | 1e-145 | 73.82% (359) | ion channel membrane protein |
| M. thermoresistible (build 8) | TH_2097 | - | 1e-135 | 70.25% (353) | POSSIBLE TRANSMEMBRANE CATION TRANSPORTER |
| M. ulcerans Agy99 | MUL_2519 | - | 1e-138 | 70.99% (355) | transmembrane cation transporter |
| M. vanbaalenii PYR-1 | Mvan_1804 | - | 1e-132 | 68.07% (357) | TrkA domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1945|M.smegmatis_MC2_155 MAKGRLRRRLAAIDSNLTSQPDAALVDVLRIPEPFISPSRKIAMRLLYAT
TH_2097|M.thermoresistible__bu -----LRRRLAAIEQDLTSQPEARLVDILRIPEPFVSPTQRIVRRIIYAT
Mflv_4663|M.gilvum_PYR-GCK MAEGRLRRRLRRFDQSLADMPVHALTDRVQIPTETVSPARRITVRVIYAL
Mvan_1804|M.vanbaalenii_PYR-1 MAEGRLRRRLSRFDQSLAEMPVHSLTDRVQIPTETVSPARRITVRVIYAL
Mb3225c|M.bovis_AF2122/97 -MAG-SWRRLRGLDEKLTAQPGYALVGVLRIPQRRASPARVISRRVVVAV
Rv3200c|M.tuberculosis_H37Rv -MAG-SWRRLRGLNEKLTAQPGYALVGVLRIPQRRASPARVISRRVVVAV
MMAR_1360|M.marinum_M MAKG-SWRRLRRLDERLTAQPSYALVGVLRIPQGQASPARIITRRVTIAL
MUL_2519|M.ulcerans_Agy99 MAKG-SWRRLRRLDERLTAQPSYALVGVLRIPQGQASPARIITRRVTIAL
MAV_4146|M.avium_104 MRNG-RSRKLSGLNQTLAAQRGHQLVGVVRIPEEHASPIRVITRRLAIAL
MAB_3514c|M.abscessus_ATCC_199 MAGK-LRQRFRGIDQALTAQPDHALVGVLRIPENAASPWRAIGKRILIAV
::: ::. *: *.. ::** ** : * *: *
MSMEG_1945|M.smegmatis_MC2_155 GALFAAVLIVYLDRHGYRDINATPDENDPLSFLDCLYYATVSLSTTGYGD
TH_2097|M.thermoresistible__bu LALMAAVVIVYLDRDGYRDVQGNE-----LSFLDCLYYATVSLSTTGYGD
Mflv_4663|M.gilvum_PYR-GCK GALFAAVLIVYLDRDGYRDSQENP-----LSFLDCLYYATVSLSTTGYGD
Mvan_1804|M.vanbaalenii_PYR-1 AALFAAVLIVYMDRDGYRDSQENQ-----LSFLDCLYYATVSLSTTGYGD
Mb3225c|M.bovis_AF2122/97 VALLLTAGIVYVDRDGYLDAQ-----GDRLTFLDCLYYAAVTLSTTGYGD
Rv3200c|M.tuberculosis_H37Rv VALLLTAGIVYVDRDGYLDAQ-----GDRLTFLDCLYYAAVTLSTTGYGD
MMAR_1360|M.marinum_M AALLAGAITVYLDRDGYHDSQ-----GDELSFLDCLYYSAVTLSTTGYGD
MUL_2519|M.ulcerans_Agy99 AALLAGAITVYLDRDGYHDSQ-----GDELSFLDCLYYSAVTLSTTGYGD
MAV_4146|M.avium_104 VVLFAAAVIVYADRSGYRDLR-----GGSLTFLDCVYFSAVSLSTTGYGD
MAB_3514c|M.abscessus_ATCC_199 GTLFAAVLVVYADRSGYRDVASTPEESDPLSFLDCFYYATVSLSTTGYGD
.*: . ** ** ** * *:****.*:::*:********
MSMEG_1945|M.smegmatis_MC2_155 ITPYTESARLVNVLVITPLRVAFLIVLIGTTVETLTTQSRQALKIQRWRS
TH_2097|M.thermoresistible__bu ITPVTPEARLINVLVITPLRVAFLIVLIGTTVETLTSQSRAALKIQRWRS
Mflv_4663|M.gilvum_PYR-GCK ITPITPEARLINVLVITPLRVAFLIVLIGTTVETLTTQSRQVYQIQRWRS
Mvan_1804|M.vanbaalenii_PYR-1 ITPITPTARLVNVLVITPLRVAFLIVLIGTTVETLTTQSRQVYQIQRWRS
Mb3225c|M.bovis_AF2122/97 ITPISEFARAINIFVITPLRIAFLILLVGTTLEVLTETSRQAYKIQRWRS
Rv3200c|M.tuberculosis_H37Rv ITPISEFARAINIFVITPLRIAFLILLVGTTLEVLTETSRQAYKIQRWRS
MMAR_1360|M.marinum_M ITPISELARAANVLIITPLRIAFLILLVGTTLEVVTETSRQALKIQRWRS
MUL_2519|M.ulcerans_Agy99 ITPISELARAANVLIITPLRIAFLILLVGTTLEVVTETSRQALKIRRWRS
MAV_4146|M.avium_104 ITPYTETARLVHTLIFTALRIAFLAVLVGTTLEVLSERSRQGWKIQRWRS
MAB_3514c|M.abscessus_ATCC_199 ITPYTEGARLVNILVITPLRLLFLIVLVGTTVEALTERSRQALKIQRWRA
*** : ** : :::*.**: ** :*:***:*.:: ** :*:***:
MSMEG_1945|M.smegmatis_MC2_155 KVRNHTVVIGYGTKGRTAVAAMVGDDVAPADIVVVDENASALERARSAGL
TH_2097|M.thermoresistible__bu RVRNHTVVIGYGTKGKTAVAAMVEDEVAPADIVVVDDDPTALERARAAGL
Mflv_4663|M.gilvum_PYR-GCK RLRNHIIIVGYGTKGRTAAAAMVGDEIAPGDIVVVDEDTSALDRAKSAGL
Mvan_1804|M.vanbaalenii_PYR-1 RLRNHIIIVGYGTKGRTAAAAMVGDEVAPADIVVVDEDAAALERAKSAGL
Mb3225c|M.bovis_AF2122/97 RVRNHTVVIGYGTKGKTAVAAMVSDELVPGEIVVVDTDSGVLERAAAAGL
Rv3200c|M.tuberculosis_H37Rv RVRNHTVVIGYGTKGKTAVAAMVSDELVPGEIVVVDTDSGVLERAAAAGL
MMAR_1360|M.marinum_M RVRNHTVVIGYGTKGKTAISAMLGDEAVPGEIVVVDTDRGALDRAAAAGL
MUL_2519|M.ulcerans_Agy99 RVRNHTVVIGYGTKGKTAISAMLGDEAVPGEIVVVDTDRGALDRAAAAGL
MAV_4146|M.avium_104 RVRNHTIVIGYGTKGKTAVAAILGDETTQAEVVVVDTDRSTLEHAESADL
MAB_3514c|M.abscessus_ATCC_199 RVRNHTVVVGYGTKGKTAVQAMLSDGAAPSEIVVVDEDQMALDAAAAADL
::*** :::******:** *:: * . .::**** : .*: * :*.*
MSMEG_1945|M.smegmatis_MC2_155 VTVHGDATKSDILRLASAQHAKAIIVATDDDASAVLVTLTARELAPKAKI
TH_2097|M.thermoresistible__bu VTVHGDATKAEVLRLASVQHAKSIIVATDNDASAVLVTLTARELAPNAKI
Mflv_4663|M.gilvum_PYR-GCK VTVRGTATDSEVLRLAGAQHAKTIIVAANRDDTAVLVTLTARELAPRAQI
Mvan_1804|M.vanbaalenii_PYR-1 VTVRGSATDSEVLRLAGAQHAKSIIVATNRDDTAVLVTLTARELAPNAKI
Mb3225c|M.bovis_AF2122/97 VTVHGDATKSDVLRLAGTQHASSIIVATSRDDTAVLVTLTAREIAPKAKI
Rv3200c|M.tuberculosis_H37Rv VTVHGDATKSDVLRLAGTQHASSIIVATSRDDTAVLVTLTAREIAPKAKI
MMAR_1360|M.marinum_M VTVHGDATKSDVLRLAGAQHAASIIVAASRDDTAALVTLTAREISPKSQI
MUL_2519|M.ulcerans_Agy99 VIVHGDATKSDVLRLAGAQHAASIIVAASRDDTAALVTLTAREISPKSQI
MAV_4146|M.avium_104 VTVHGDATKADVLRLAGAQHAASIIVATSRDDTAVLVTLTAREIAPHAKI
MAB_3514c|M.abscessus_ATCC_199 VTVRGSATKSDVLRLAGVQNAKSIIVAANRDDTSVLVTLTARELAPNAKI
* *:* **.:::****..*:* :****:. * ::.********::*.::*
MSMEG_1945|M.smegmatis_MC2_155 IAAAREAENQHLLRQSGADSTVVSSETAGRLLGIATQTPSVVEMMEDLLT
TH_2097|M.thermoresistible__bu IAAARESDNQHLLKQSGADSTVVSSETAGRLLGVATQTPSVVAIIEDLLT
Mflv_4663|M.gilvum_PYR-GCK IAAVRETDNEHLLKQSGADSTVVTSETAGRLLGIAVQTPSVVEMMEDLLT
Mvan_1804|M.vanbaalenii_PYR-1 IAAVREAENQHLLEQSGADSTVVTSETAGRLLGIAVQTPSVVEMMEDLLT
Mb3225c|M.bovis_AF2122/97 VASIREAENQHLLRQSGADTVVVSSETAGRLLGIATTTPSVVEMIEDLLT
Rv3200c|M.tuberculosis_H37Rv VASIREAENQHLLRQSGADTVVVSSETAGRLLGIATTTPSVVEMIEDLLT
MMAR_1360|M.marinum_M VASIREAENQHLLEQSGADSVVVSSETAGRLLGLATTTPSVVQMIEDLLT
MUL_2519|M.ulcerans_Agy99 VASIREAENQHLLEQSGADSVVVSSETAGRLLGLATTTPSVVQMIEDLLT
MAV_4146|M.avium_104 VASIREAENQHLLQQSGADSVVVSSATAGRLLGLATTTPSVVEMIEDLLT
MAB_3514c|M.abscessus_ATCC_199 VAAIREAENVHLLRQSGADSVVVSSETAGRLLGIATTTPRVVEMIEDLLT
:*: **::* ***.*****:.**:* *******:*. ** ** ::*****
MSMEG_1945|M.smegmatis_MC2_155 PDAGFAIAEREVSPKEEGGSPRHLHDIVLGVVRGGRLVRVDDPEVDALEA
TH_2097|M.thermoresistible__bu PDAGFSIAEREVTPKEVGGSPRHLQDIVLGVVRDGHLYRVDEPEVDAVEA
Mflv_4663|M.gilvum_PYR-GCK PDRGFAIAEREVSPKEAGGSPRHQTDIVLGVVRNKKLIRVDDPEVDALEL
Mvan_1804|M.vanbaalenii_PYR-1 PDRGFAIAEREVSPKEAGSSPRHQTDIVLGVVRDNKLIRVDEPEVDALEL
Mb3225c|M.bovis_AF2122/97 PEAGLAVAEREVEQAEVGGSPRHLRDIVLGVVRDGQLLRIGAPEVDAIEA
Rv3200c|M.tuberculosis_H37Rv PEAGLAVAEREVEQAEVGGSPRHLRDIVLGVVRDGQLLRIGAPEVDAIEA
MMAR_1360|M.marinum_M PDAGLAIAEREVEQSEIGGSPRHLRDIVLGVVRGGQLLRIGAPEVDAIEA
MUL_2519|M.ulcerans_Agy99 PDAGLAIAEREVEQSEIGGSPRHLRDIVLGVVRGGQLLRIGAPEVDAIEA
MAV_4146|M.avium_104 PDVGLAIAEREVEQSEIGGSPRHLRDIVLGVVRDGRLLRIDAPEVDAVEA
MAB_3514c|M.abscessus_ATCC_199 PEAGFAIAEREVERSEVGGSPRHLSDIVLGVVRDGILHRVDAPEVDSIEA
*: *:::***** * *.**** ********. * *:. ****::*
MSMEG_1945|M.smegmatis_MC2_155 GDRLLYIRSADAER----------------------------------
TH_2097|M.thermoresistible__bu GDRLLYIRISDGDR----------------------------------
Mflv_4663|M.gilvum_PYR-GCK GDRLLYIRSADEDR----------------------------------
Mvan_1804|M.vanbaalenii_PYR-1 GDRLLYIRSADEER----------------------------------
Mb3225c|M.bovis_AF2122/97 SDRLLYIRQVGR------------------------------------
Rv3200c|M.tuberculosis_H37Rv SDRLLYIRQVGR------------------------------------
MMAR_1360|M.marinum_M NDRLLYIRNTGH------------------------------------
MUL_2519|M.ulcerans_Agy99 NDRLLYIRNTGALVAIASAAQPGAAGRHQGGGVAIVSSAQPGCQGGES
MAV_4146|M.avium_104 NDRLLYIRSAGH------------------------------------
MAB_3514c|M.abscessus_ATCC_199 TDRLLYVRNAGA------------------------------------
*****:* .