For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AEFIYTMKKVRKAHGDKVILDDVTLSFYPGAKIGVVGPNGAGKSSVLRIMAGLDKPNNGDAFLATGATVG ILLQEPPLNEEKTVRGNVEEGLGDIKVKLDRFNEVAELMATDYSDELMEEMGRLQEDLDHADAWDIDSQL EQAMDALRCPPPEEPVANLSGGERRRVALCKLLLSKPDLLLLDEPTNHLDAESVQWLEQHLASYAGAVLA VTHDRYFLDNVAEWILELDRGRAYPYEGNYSTYLEKKADRLAVQGRKDAKLQKRLQEELAWVRSGAKARQ AKSKARLQRYEEMVAEAEKTRKLDFEEIQIPVGPRLGSVVVEVEHLDKGFGGRTLIKDLSFTLPRNGIVG VIGPNGVGKTTLFKTIVGLEEPDSGTVKVGETVKLSYVDQTRAGIDPKKTVWEVVSDGLDYIEVGQSEVP SRAYVSAFGFKGPDQQKPAGVLSGGERNRLNLALTLKQGGNLILLDEPTNDLDVETLSSLENALVNFPGC AVVISHDRWFLDRTCTHILAWEGDADSEAKWFWFEGNFGAYEENKVERLGVEAARPHRVTHRRLTRD*
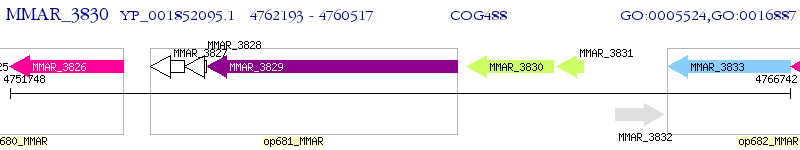
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3830 | - | - | 100% (558) | ATP-binding component of an ABC transporter |
| M. marinum M | MMAR_2476 | - | 6e-70 | 33.66% (505) | ABC transporter, ATP-binding protein |
| M. marinum M | MMAR_2279 | - | 3e-63 | 32.87% (505) | ATP-binding protein ABC transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2504c | - | 0.0 | 93.90% (557) | putative ABC transporter ATP-binding protein |
| M. gilvum PYR-GCK | Mflv_2568 | - | 0.0 | 90.32% (558) | putative ABC transporter ATP-binding protein |
| M. tuberculosis H37Rv | Rv2477c | - | 0.0 | 93.90% (557) | putative ABC transporter ATP-binding protein |
| M. leprae Br4923 | MLBr_01248 | - | 0.0 | 92.11% (558) | putative ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_1560 | - | 0.0 | 89.43% (558) | putative ABC transporter ATP-binding protein |
| M. avium 104 | MAV_1695 | - | 0.0 | 95.52% (558) | putative ABC transporter ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_4700 | - | 0.0 | 90.86% (558) | putative ABC transporter ATP-binding protein |
| M. thermoresistible (build 8) | TH_1035 | - | 0.0 | 91.04% (558) | PROBABLE MACROLIDE-TRANSPORT ATP-BINDING PROTEIN ABC |
| M. ulcerans Agy99 | MUL_3757 | - | 0.0 | 99.28% (558) | putative ABC transporter ATP-binding protein |
| M. vanbaalenii PYR-1 | Mvan_4070 | - | 0.0 | 91.02% (557) | putative ABC transporter ATP-binding protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2504c|M.bovis_AF2122/97 MAEFIYTMKKVRKAHGDKVILDDVTLSFYPGAKIGVVGPNGAGKSSVLRI
Rv2477c|M.tuberculosis_H37Rv MAEFIYTMKKVRKAHGDKVILDDVTLSFYPGAKIGVVGPNGAGKSSVLRI
MLBr_01248|M.leprae_Br4923 MAEFIYTMKKVRKAHGDKVILDDVTLNFFPGAKIGVVGPNGAGKSSVLRI
MMAR_3830|M.marinum_M MAEFIYTMKKVRKAHGDKVILDDVTLSFYPGAKIGVVGPNGAGKSSVLRI
MUL_3757|M.ulcerans_Agy99 MAEFIYTMKKVRKAHGDKVILDDVTLSFYPGAKIGVVGPNGAGKSSVLRI
MAV_1695|M.avium_104 MAEFIYTMKKVRKAHGDKVILDDVTLSFYPGAKIGVVGPNGAGKSSVLRI
TH_1035|M.thermoresistible__bu MAEFIYTMRKVRKAHGDKVILDDVSLNFLPGAKIGVVGPNGAGKSSVLRI
Mflv_2568|M.gilvum_PYR-GCK MAEFIYTMRKVRKAHGDKVILDDVTLNFLPGAKIGVVGPNGAGKSSVLRI
Mvan_4070|M.vanbaalenii_PYR-1 MAEFIYTMRKVRKAHGDKVILDDVTLNFLPGAKIGVVGPNGAGKSSVLRI
MSMEG_4700|M.smegmatis_MC2_155 MAEFIYTMRKVRKAHGDKVILDDVTLNFLPGAKIGVVGPNGAGKSSVLRI
MAB_1560|M.abscessus_ATCC_1997 MAEYIYTMMKVRKAHGDKVILDDVTLNFLPGAKIGVVGPNGAGKSSVLKI
***:**** ***************:*.* *******************:*
Mb2504c|M.bovis_AF2122/97 MAGLDKPNNGDAFLATGATVGILQQEPPLNEDKTVRGNVEEGMGDIKIKL
Rv2477c|M.tuberculosis_H37Rv MAGLDKPNNGDAFLATGATVGILQQEPPLNEDKTVRGNVEEGMGDIKIKL
MLBr_01248|M.leprae_Br4923 MAGMDKPNNGDAFLATGASVGILQQEPPLNEEKTVRGNVEEGLGDIKIKL
MMAR_3830|M.marinum_M MAGLDKPNNGDAFLATGATVGILLQEPPLNEEKTVRGNVEEGLGDIKVKL
MUL_3757|M.ulcerans_Agy99 IAGLDKPNNGDAFLATGATVGILLQEPPLNEEKTVRGNVEEGLGDIKVKL
MAV_1695|M.avium_104 MAGLDKPNNGEAFLATDATVGILQQEPPLNEEKTVRGNVEEGLGEIKVKL
TH_1035|M.thermoresistible__bu MAGLDQPNNGEAFLQPGATVGILLQEPPLNEEKTVRGNVEEGLGEIKVKL
Mflv_2568|M.gilvum_PYR-GCK MAGLDQPNNGDAFLQPGATVGILMQEPQLDETKTVRENVEDGV-AIKGKL
Mvan_4070|M.vanbaalenii_PYR-1 MAGLDQPNNGDAFLAPGATVGILLQEPELDETKTVRENVEEGV-AIKAKL
MSMEG_4700|M.smegmatis_MC2_155 MAGLDQPNNGDAFLAPDATVGILMQEPQLDETKTVRENVEDGV-AIKGKL
MAB_1560|M.abscessus_ATCC_1997 MAGLDKPNNGEAFLANGASVGILQQEPPLDETKTVRENVEDGV-AVKAKL
:**:*:****:*** .*:**** *** *:* **** ***:*: :* **
Mb2504c|M.bovis_AF2122/97 DRFNEVAELMATDYTDELMEEMGRLQEELDHADAWDLDAQLEQAMDALRC
Rv2477c|M.tuberculosis_H37Rv DRFNEVAELMATDYTDELMEEMGRLQEELDHADAWDLDAQLEQAMDALRC
MLBr_01248|M.leprae_Br4923 DRFSEVAELMATDSSSELMEEMGRLQEELDHADAWDLDSQLEQAMDVLRC
MMAR_3830|M.marinum_M DRFNEVAELMATDYSDELMEEMGRLQEDLDHADAWDIDSQLEQAMDALRC
MUL_3757|M.ulcerans_Agy99 DRFNEVAELMATDYSDELMEEMGRLQEDLDHADAWDIDSQLEQAMDALRC
MAV_1695|M.avium_104 DRFNEVAELMATDYSDELMEEMGRLQEELDHADAWDLDSQLEQAMDALRC
TH_1035|M.thermoresistible__bu DRFNEVAELMATDYSDELMEEMGRLQEELDHADAWDLDSQLEQAMDALRC
Mflv_2568|M.gilvum_PYR-GCK NRYNEVAELMATDYTDELMDEMGKLQEELDAADAWDIDSQLEQAMDALRC
Mvan_4070|M.vanbaalenii_PYR-1 NRYNEVAELMATDYTDELMEEMGKLQEELDAADAWDIDSQLEQAMDALRC
MSMEG_4700|M.smegmatis_MC2_155 NRYNEVAELMATDYSDELMEEMGKLQEELDAADAWDIDSQLEQAMDALRC
MAB_1560|M.abscessus_ATCC_1997 NRYNEVAELMATDYSDELMEEMGKLQEELDAADAWDIDSQLEQAMDALRC
:*:.********* :.***:***:***:** *****:*:*******.***
Mb2504c|M.bovis_AF2122/97 PPADEPVTNLSGGERRRVALCKLLLSKPDLLLLDEPTNHLDAESVQWLEQ
Rv2477c|M.tuberculosis_H37Rv PPADEPVTNLSGGERRRVALCKLLLSKPDLLLLDEPTNHLDAESVQWLEQ
MLBr_01248|M.leprae_Br4923 PPPDEPVTNLSGGERRRVALCKLLLSKPDLLLLDEPTNHLDAESVQWLEQ
MMAR_3830|M.marinum_M PPPEEPVANLSGGERRRVALCKLLLSKPDLLLLDEPTNHLDAESVQWLEQ
MUL_3757|M.ulcerans_Agy99 PPPEEPVANLSGGERRRVALCKLLLSKPDLLLLDEPTNHLDAESVQWLEQ
MAV_1695|M.avium_104 PPPDDPVTNLSGGERRRVALCKLLLSKPDLLLLDEPTNHLDAESVQWLEQ
TH_1035|M.thermoresistible__bu PPPDEPVTHLSGGERRRVALCKLLLSKPDLLLLDEPTNHLDAESVLWLEQ
Mflv_2568|M.gilvum_PYR-GCK PPPDEPVTHLSGGEKRRVALCKLLLSKPDLLLLDEPTNHLDAESVLWLEQ
Mvan_4070|M.vanbaalenii_PYR-1 PPPDEPVTHLSGGEKRRVALCKLLLSKPDLLLLDEPTNHLDAESVLWLEQ
MSMEG_4700|M.smegmatis_MC2_155 PPPDEPVTHLSGGERRRVALCKLLLSKPDLLLLDEPTNHLDAESVLWLEQ
MAB_1560|M.abscessus_ATCC_1997 PPPDEPVTHLSGGERRRVALCKLLLSKPDLLLLDEPTNHLDAESVLWLEQ
**.::**::*****:****************************** ****
Mb2504c|M.bovis_AF2122/97 HLASYPGAILAVTHDRYFLDNVAEWILELDRGRAYPYEGNYSTYLEKKAE
Rv2477c|M.tuberculosis_H37Rv HLASYPGAILAVTHDRYFLDNVAEWILELDRGRAYPYEGNYSTYLEKKAE
MLBr_01248|M.leprae_Br4923 HLAGYAGAVLAVTHDRYFLDNVAEWILELDRGRAYPYEGNYSTYLEKKAE
MMAR_3830|M.marinum_M HLASYAGAVLAVTHDRYFLDNVAEWILELDRGRAYPYEGNYSTYLEKKAD
MUL_3757|M.ulcerans_Agy99 HLASYAGAVLAVTHDRYFLDNVAEWILELDRGRTYPYEGNYSTYLEKKAD
MAV_1695|M.avium_104 HLAAYAGAVLAVTHDRYFLDNVAQWILELDRGRAYPYEGNYSTYLEKKAD
TH_1035|M.thermoresistible__bu HLAEYKGAVLAVTHDRYFLDNVAEWILELDRGRAYPYEGNYSTYLEKKAE
Mflv_2568|M.gilvum_PYR-GCK HLAEYKGAILAVTHDRYFLDNVAEWILELDRGRAYPYEGNYSTYLEKKAE
Mvan_4070|M.vanbaalenii_PYR-1 HLAEYKGAVLAVTHDRYFLDNVAEWILELDRGRAFPYEGNYSTYLEKKAE
MSMEG_4700|M.smegmatis_MC2_155 HLASYPGAILAVTHDRYFLDNVAEWILELDRGRAYPYEGNYSTYLEKKAE
MAB_1560|M.abscessus_ATCC_1997 HLATYPGAILAVTHDRYFLDNVAEWILELDRGRAYPYEGNYSTYLEKKAE
*** * **:**************:*********::**************:
Mb2504c|M.bovis_AF2122/97 RLAVQGRKDAKLQKRLTEELAWVRSGAKARQAKSKARLQRYEEMAAEAEK
Rv2477c|M.tuberculosis_H37Rv RLAVQGRKDAKLQKRLTEELAWVRSGAKARQAKSKARLQRYEEMAAEAEK
MLBr_01248|M.leprae_Br4923 RLTTQGRKDAKLQKRLTDELAWVRSGAKARQAKSKARLQRYEEMAAEAEK
MMAR_3830|M.marinum_M RLAVQGRKDAKLQKRLQEELAWVRSGAKARQAKSKARLQRYEEMVAEAEK
MUL_3757|M.ulcerans_Agy99 RLAVQGRKDAKLQKRLQEELAWVRSGAKARQAKSKARLQRYEEMVAEAEK
MAV_1695|M.avium_104 RLAVQGRKDAKLQKRLSEELAWVRSGAKARQAKSKARLQRYEEMAAEAEK
TH_1035|M.thermoresistible__bu RLEVQGRKDAKLHKRLKEELAWVRSGTKARQAKGKARLQRYEEMAAEAEK
Mflv_2568|M.gilvum_PYR-GCK RIAVTGRKDAKLQKRLKDELEWVRSGAKARQAKNKARLQRYEEMAAEAEK
Mvan_4070|M.vanbaalenii_PYR-1 RIAVQGRKDAKLQKRLKDELDWVRSGAKARQAKSKARLQRYEEMAAEAEK
MSMEG_4700|M.smegmatis_MC2_155 RLEVQGKKDQKLQKRLKEELAWVRSGAKARQAKNKARLDRYEEMVAEAEK
MAB_1560|M.abscessus_ATCC_1997 RIAVQGRKDAKLHKRLQEELAWVRSGAKARQAKNKARLQRYEEMVTEAEK
*: . *:** **:*** :** *****:******.****:*****.:****
Mb2504c|M.bovis_AF2122/97 TRKLDFEEIQIPVGPRLGNVVVEVDHLDKGYDGRALIKDLSFSLPRNGIV
Rv2477c|M.tuberculosis_H37Rv TRKLDFEEIQIPVGPRLGNVVVEVDHLDKGYDGRALIKDLSFSLPRNGIV
MLBr_01248|M.leprae_Br4923 NRKFDFEEIQIPVGPRLGNMVVEVEHLDKGYGGRTLIKDLSFTLPRNGIV
MMAR_3830|M.marinum_M TRKLDFEEIQIPVGPRLGSVVVEVEHLDKGFGGRTLIKDLSFTLPRNGIV
MUL_3757|M.ulcerans_Agy99 TRKLDFEEIQIPVGPRLGSVVVEVEHLDKGFGGRTLIKDLSFTLPRNGTV
MAV_1695|M.avium_104 TRKLDFEEIQIPVGPRLGNVVVEVEHLDKGYDGRTLIKDLSFTLPRNGIV
TH_1035|M.thermoresistible__bu SRKLDFEEIQIPTPPRLGNVVVEVKNLDKGYDGRPLIKDLSFTLPRNGIV
Mflv_2568|M.gilvum_PYR-GCK TRKLDFEEIQIPVGPRLGNVVVEVEHLDKGFDGRLLIKDLSFTLPRNGIV
Mvan_4070|M.vanbaalenii_PYR-1 TRKLDFEEIQIPTGPRLGNVVVEVEHLDKGFGGRQLIKDLSFTLPRNGIV
MSMEG_4700|M.smegmatis_MC2_155 TRKLDFEEIQIPTPPRLGSVVVEVEHLDKGFNGRTLIKDLSFTLPRNGIV
MAB_1560|M.abscessus_ATCC_1997 TRKLDFEEIQIPTGPRLGNVVVEVEHLDKGFEGRALIKDLSFTLPRNGIV
.**:********. ****.:****.:****: ** *******:***** *
Mb2504c|M.bovis_AF2122/97 GVIGPNGVGKTTLFKTIVGLETPDSGSVKVGETVKLSYVDQARAGIDPRK
Rv2477c|M.tuberculosis_H37Rv GVIGPNGVGKTTLFKTIVGLETPDSGSVKVGETVKLSYVDQARAGIDPRK
MLBr_01248|M.leprae_Br4923 GIIGPNGVGKTTLFKTIVGLEQPDGGAVKIGETVKLSYVDQTRAGIDPKK
MMAR_3830|M.marinum_M GVIGPNGVGKTTLFKTIVGLEEPDSGTVKVGETVKLSYVDQTRAGIDPKK
MUL_3757|M.ulcerans_Agy99 GVIGPNGVGKTTLFKTIVGLEEPDSGTVKVGETVKLSYVDQTRAGIDPKK
MAV_1695|M.avium_104 GVIGPNGVGKTTLFKTIVGLEQPDSGTVKVGETVKLSYVDQARAGIDPKK
TH_1035|M.thermoresistible__bu GVIGPNGVGKTTLFKTIVGLEEPDSGTVKIGETVKLSYVDQTRAGIDPNK
Mflv_2568|M.gilvum_PYR-GCK GVIGPNGVGKTTLFKTIVGLEEPDSGTVKVGETVKLSYVDQTRGGIDPKK
Mvan_4070|M.vanbaalenii_PYR-1 GVIGPNGVGKTTLFKTIVGLEQPDSGTVKIGETVKLSYVDQSRAGIDPKK
MSMEG_4700|M.smegmatis_MC2_155 GVIGPNGVGKTTLFKTIVGLEQPDSGTVKIGETVKLSYVDQTRAGIDPKK
MAB_1560|M.abscessus_ATCC_1997 GVIGPNGVGKTTLFKTIVGLENPDGGEVKVGETVKLSYVDQSRAGIDPKK
*:******************* **.* **:***********:*.****.*
Mb2504c|M.bovis_AF2122/97 TVWEVVSDGLDYIQVGQTEVPSRAYVSAFGFKGPDQQKPAGVLSGGERNR
Rv2477c|M.tuberculosis_H37Rv TVWEVVSDGLDYIQVGQTEVPSRAYVSAFGFKGPDQQKPAGVLSGGERNR
MLBr_01248|M.leprae_Br4923 IVWEVVSDGLDHIQVGQTEVSSRAYVSAFGFKGPDQQKLAGVLSGGERNR
MMAR_3830|M.marinum_M TVWEVVSDGLDYIEVGQSEVPSRAYVSAFGFKGPDQQKPAGVLSGGERNR
MUL_3757|M.ulcerans_Agy99 TVWEVVSDGLDSIEVGQSEVPSRAYVSAFGFKGPDQQKPAGVLSGGERNR
MAV_1695|M.avium_104 TVWEVVSDGLDYIEVGQNEVPSRAYVSAFGFKGPDQQKPAGVLSGGERNR
TH_1035|M.thermoresistible__bu TVWEVVSDGLDYIEVGQNEIPSRAYVSAFGFKGPDQQKPAGVLSGGERNR
Mflv_2568|M.gilvum_PYR-GCK TVWEVVSDGLDFIEVGQNEVPSRAYVSAFGFKGPDQQKPAGVLSGGERNR
Mvan_4070|M.vanbaalenii_PYR-1 TVWEVVSDGLDYIEVGQNEIPSRAYVSAFGFKGPDQQKPAGVLSGGERNR
MSMEG_4700|M.smegmatis_MC2_155 TVWQVVSDGLDYIEVGQNEIPSRAYVSAFGFKGPDQQKPAGVLSGGERNR
MAB_1560|M.abscessus_ATCC_1997 NVWEVVSDGLDHIVVGQNEMPSRAYVSAFGFKGPDQQKPAGVLSGGERNR
**:******* * ***.*:.***************** ***********
Mb2504c|M.bovis_AF2122/97 LNLALTLKQGGNLILLDEPTNDLDVETLGSLENALLNFPGCAVVISHDRW
Rv2477c|M.tuberculosis_H37Rv LNLALTLKQGGNLILLDEPTNDLDVETLGSLENALLNFPGCAVVISHDRW
MLBr_01248|M.leprae_Br4923 LNLALTLKQGGNLILLDEPTNDLDVETLGSLENALVNFPGCAVVISHDRW
MMAR_3830|M.marinum_M LNLALTLKQGGNLILLDEPTNDLDVETLSSLENALVNFPGCAVVISHDRW
MUL_3757|M.ulcerans_Agy99 LNLALTLKQGGNLILLDEPTNDLDVETLSSLENALVNFPGCAVVISHDRW
MAV_1695|M.avium_104 LNLALTLKQGGNLILLDEPTNDLDVETLGSLENALVNFPGCAVVISHDRW
TH_1035|M.thermoresistible__bu LNLALTLKEGGNLILLDEPTNDLDVETLGSLENALEKFPGCAVVISHDRW
Mflv_2568|M.gilvum_PYR-GCK LNLALTLKEGGNLILLDEPTNDLDVETLSSLENALQNFPGCAVVISHDRW
Mvan_4070|M.vanbaalenii_PYR-1 LNLALTLKEGGNLLLLDEPTNDLDVETLSSLENALEKFPGCAVVISHDRW
MSMEG_4700|M.smegmatis_MC2_155 LNLALTLKEGGNLILLDEPTNDLDVETLSSLENALQQFPGCAVVISHDRW
MAB_1560|M.abscessus_ATCC_1997 LNLALTLKQGGNLILLDEPTNDLDVETLGSLENALEQFPGCAVVISHDRW
********:****:**************.****** :*************
Mb2504c|M.bovis_AF2122/97 FLDRTCTHILAWEGDDDNEAKWFWFEGNFGAYEENKVERLGVDAARPHRV
Rv2477c|M.tuberculosis_H37Rv FLDRTCTHILAWEGDDDNEAKWFWFEGNFGAYEENKVERLGVDAARPHRV
MLBr_01248|M.leprae_Br4923 FLDRTCTHILAWEGDD--EGKWFWFEGNFGAYEENKVERLGAEAARPHRV
MMAR_3830|M.marinum_M FLDRTCTHILAWEGDADSEAKWFWFEGNFGAYEENKVERLGVEAARPHRV
MUL_3757|M.ulcerans_Agy99 FLDRTCTHILAWEGDADSEAKWFWFEGNFGAYEENKVERLGVEAARPHRV
MAV_1695|M.avium_104 FLDRTCTHILAWEGDDDNEAKWFWFEGNFGAYEENKIERLGAEAARPHRV
TH_1035|M.thermoresistible__bu FLDRVCTHILAWEGTEDNPAQWFWFEGNFAAYEENKVARLGPEAARPHRV
Mflv_2568|M.gilvum_PYR-GCK FLDRTCTHILAWEGDSDNEAKWFWFEGNFGGYEENKIERLGADAARPHRV
Mvan_4070|M.vanbaalenii_PYR-1 FLDRTCTHILAWEGDADNEAKWFWFEGNFGGYEENKIERLGAEAARPHRV
MSMEG_4700|M.smegmatis_MC2_155 FLDRTCTHILAWEGDDDNEAKWFWFEGNFGAYEENKIQRMGAEAARPHRV
MAB_1560|M.abscessus_ATCC_1997 FLDRTCTHILAWEGSDDNPASWFWFEGNFQAYEENKVERLGIDAARPHRV
****.********* ..******** .*****: *:* :*******
Mb2504c|M.bovis_AF2122/97 THRKLTRG
Rv2477c|M.tuberculosis_H37Rv THRKLTRG
MLBr_01248|M.leprae_Br4923 THRKLTRD
MMAR_3830|M.marinum_M THRRLTRD
MUL_3757|M.ulcerans_Agy99 THRRLTRD
MAV_1695|M.avium_104 THRKLTRD
TH_1035|M.thermoresistible__bu THRRLTRD
Mflv_2568|M.gilvum_PYR-GCK THRKLTRD
Mvan_4070|M.vanbaalenii_PYR-1 THRKLTRG
MSMEG_4700|M.smegmatis_MC2_155 THRRLTRD
MAB_1560|M.abscessus_ATCC_1997 THRRLTRD
***:***.