For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVGASGFASGAVSGSGGPRLPTLTDLLYQLASGSVTSVELTRRSLREIEVSQSTLNAFRVVLTESALADA AAADRRRAAGDAAPLLGIPIAVKDDVDVAGVPTAFGTQGYVRPAAHDSEVVRRLKAAGAVIIGKTNTCEL GQWPFTSGPGFGHTRNPWARRHTPGGSSGGSAAAVAAGLVTAAIGSDGAGSIRVPAAWTHLVGIKPQRGR ISTWPWPETFNGITVNGVLARTVADAALVLDAASGNVEGDLHKPAPLTASDYVGIAPGPLKIAMSTRFPY TGFRAKLHPEILAATRRVGEQLQLLGHTVERGNPDYGLRLSWDFLARSTAGLWEWAECLGDGVTLDPRTV TNMRAGHALSQAILRNARHHEAAAQRRVGSIFDIVDVVLAPTTAQPPPLARAFDDMGGLATDRAIIAACP LTWPWNLLGWPSINVPAGFTVDGLPIGVQLMGPAESEGLLISLAAELEAVSGWATKQPKVWWNTGTSTPP IHSVRPPRR
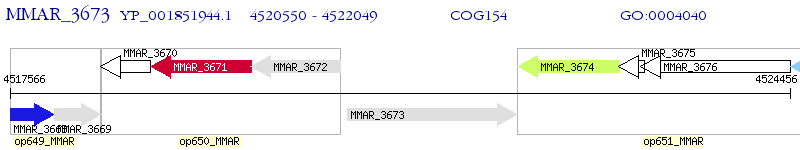
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3673 | amiA2 | - | 100% (499) | amidase AmiA2 |
| M. marinum M | MMAR_4176 | amiB2 | 2e-93 | 44.91% (452) | amidase AmiB2 |
| M. marinum M | MMAR_3565 | amiC_1 | 5e-53 | 34.78% (483) | amidase AmiC_1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2384 | amiA2 | 0.0 | 86.78% (484) | amidase |
| M. gilvum PYR-GCK | Mflv_2712 | - | 0.0 | 73.47% (475) | amidase |
| M. tuberculosis H37Rv | Rv2363 | amiA2 | 0.0 | 86.78% (484) | amidase |
| M. leprae Br4923 | MLBr_01702 | gatA | 7e-45 | 31.30% (492) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. abscessus ATCC 19977 | MAB_1674c | - | 1e-178 | 66.67% (462) | amidase |
| M. avium 104 | MAV_2032 | - | 0.0 | 85.71% (448) | amidase |
| M. smegmatis MC2 155 | MSMEG_4492 | - | 0.0 | 74.53% (479) | amidase |
| M. thermoresistible (build 8) | TH_4482 | - | 0.0 | 74.52% (471) | PUTATIVE Amidase |
| M. ulcerans Agy99 | MUL_3616 | amiA2 | 0.0 | 99.20% (499) | amidase |
| M. vanbaalenii PYR-1 | Mvan_3824 | - | 0.0 | 72.42% (475) | amidase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2712|M.gilvum_PYR-GCK -MPDSS-ARSTSRNRFPDRFPTVANQLFQLASGTATSVDLVRRSLVAIEA
Mvan_3824|M.vanbaalenii_PYR-1 -MADTAYSRITSRR----RFPTLGNQLFQLASGTATSVDLVSRSLQAIEA
TH_4482|M.thermoresistible__bu -MVDAYKPPAK--------LPTLTDLLYQLASGAVTSQELVRRSLRAIEA
MMAR_3673|M.marinum_M MVGASGFASGAVSGSGGPRLPTLTDLLYQLASGSVTSVELTRRSLREIEV
MUL_3616|M.ulcerans_Agy99 MVGASGFASGAVSGSGGPRLPTLTDLLYQLASGSVTSVELTRRSLREIEV
Mb2384|M.bovis_AF2122/97 MVGASGSDAGAISGSGNQRLPTLTDLLYQLATRAVTSEELVRRSLRAIDV
Rv2363|M.tuberculosis_H37Rv MVGASGSDAGAISGSGNQRLPTLTDLLYQLATRAVTSEELVRRSLRAIDV
MAV_2032|M.avium_104 ----------------------------------------MGRSLHAIHA
MSMEG_4492|M.smegmatis_MC2_155 MMGKS--------TACESTFPTLTNQLYQLASGAVTSDELVRRSLHAINA
MAB_1674c|M.abscessus_ATCC_199 ----------------MSGFPTLTSQAAALSDGSESSVSLTLTALNAIKS
MLBr_01702|M.leprae_Br4923 ----------MIGSLHEIIRLDASTLAAKIVAKELSSVEITQACLDQIEA
.* *.
Mflv_2712|M.gilvum_PYR-GCK SQPTLNAFRVVLAERALADAAEADRLRAAGHDRARRPLLGIPIAVKDDVD
Mvan_3824|M.vanbaalenii_PYR-1 SQPTLNAFRVVLTEQALADAAEADRKRAAGADLSRHPLLGVPIAVKDDVD
TH_4482|M.thermoresistible__bu SQSTLNAFRVVLADRALAEAAEADRKRAAG---EHLPLLGVPIAVKDDVD
MMAR_3673|M.marinum_M SQSTLNAFRVVLTESALADAAAADRRRAAG---DAAPLLGIPIAVKDDVD
MUL_3616|M.ulcerans_Agy99 SQSTLNAFRVVLTESALADAAAADRRRAAG---DAAPLLGIPIAVKDDVD
Mb2384|M.bovis_AF2122/97 SQPTLNAFRVVLTESALADAAAADKRRAAG---DTAPLLGIPIAVKDDVD
Rv2363|M.tuberculosis_H37Rv SQPTLNAFRVVLTESALADAAAADKRRAAG---DTAPLLGIPIAVKDDVD
MAV_2032|M.avium_104 SQPTLNAFRVVLTESALADAAEADRRRAAG---DTAPLLGIPIAVKDDVD
MSMEG_4492|M.smegmatis_MC2_155 SQSTLNAFRVVLTEQALADAAKADRDRAAG---KQLPLLGVPIAVKDDVD
MAB_1674c|M.abscessus_ATCC_199 SQSTLNAFRVVCTKTALAEATEADKRLARG---ERLPLLGVPIAIKDDTD
MLBr_01702|M.leprae_Br4923 TDDTYRAFLHVAAEKALSAAAAVDKAVAAG-GQLSSTLAGVPLALKDVFT
:: * .** * :. **: *: .*: * * .* *:*:*:**
Mflv_2712|M.gilvum_PYR-GCK VAGVPTRFGASGEV-PEAVADAEVVRRLRAAGAVIVGKTNTCELGQWPFT
Mvan_3824|M.vanbaalenii_PYR-1 VAGVPTRFGADGEV-PPAEADAEVVRRLRAAGAVIVGKTNTCELGQWPFT
TH_4482|M.thermoresistible__bu IAGVPTRFGAVGDV-PPATEDAEVVRRLRAAGAVIVGKTHTCELGQWPFT
MMAR_3673|M.marinum_M VAGVPTAFGTQGYV-RPAAHDSEVVRRLKAAGAVIIGKTNTCELGQWPFT
MUL_3616|M.ulcerans_Agy99 VAGVPTAFGTQGYV-QPAAHDSEVVRRLKAAGAVIIGKTNTCELGQWPFT
Mb2384|M.bovis_AF2122/97 VAGVPTAFGTQGYV-APATDDCEVVRRLKAAGAVIVGKTNTCELGQWPFT
Rv2363|M.tuberculosis_H37Rv VAGVPTAFGTQGYV-APATDDCEVVRRLKAAGAVIVGKTNTCELGQWPFT
MAV_2032|M.avium_104 IAGVPTAFGTEGSV-PPATHDAEVVRRLKAAGAVIVGKTNTCELGQWPFT
MSMEG_4492|M.smegmatis_MC2_155 IAGVPTRFGTEGDG-RVAEADSEVARRLRAAGAVIVGKTNTCELGQWHFT
MAB_1674c|M.abscessus_ATCC_199 LAGTPTAFGTAGAV-GSESEDAELVRRLRAAGAVIVGKTNTCELGQWGFT
MLBr_01702|M.leprae_Br4923 TVDMPTTCGSKILQGWHSPYDATVTTRLRAAGIPILGKTNMDEFAMGSST
.. ** *: *. :. **:*** *:***: *:. *
Mflv_2712|M.gilvum_PYR-GCK SGPAFGHTRNPWSREHTPGGSSGGSAAAVAAGLVAAAIGSDGAGSVRIPA
Mvan_3824|M.vanbaalenii_PYR-1 SGPAFGHTRNPWSREHTPGGSSGGSAAAVAAGLVAAAIGSDGAGSVRIPA
TH_4482|M.thermoresistible__bu SGPAFGHTRNPWSPRHTPGGSSGGSAAAVAAGLVTAAIGSDGAGSVRIPA
MMAR_3673|M.marinum_M SGPGFGHTRNPWARRHTPGGSSGGSAAAVAAGLVTAAIGSDGAGSIRVPA
MUL_3616|M.ulcerans_Agy99 SGPGFGHTRNPWARRHTPGGSSGGSAAAVAAGLVTAAIGSDGAGSIRVPA
Mb2384|M.bovis_AF2122/97 SGPGFGHTRNPWSRRHTPGGSSGGSAAAVAAGLVTAAIGSDGAGSIRIPA
Rv2363|M.tuberculosis_H37Rv SGPGFGHTRNPWSRRHTPGGSSGGSAAAVAAGLVTAAIGSDGAGSIRIPA
MAV_2032|M.avium_104 SGPGFGHTRNPWSRRHTPGGSSGGSAAAVAAGLVTAAIGSDGAGSIRIPA
MSMEG_4492|M.smegmatis_MC2_155 SGPGFGHTRNPWSRKHTPGGSSGGSAAAVAAGLVTAAIGSDGAGSVRIPA
MAB_1674c|M.abscessus_ATCC_199 SGPGFGHTRNPWSRGHTPGGSSGGSAAAVAAGLVAGAIGSDGAGSVRIPA
MLBr_01702|M.leprae_Br4923 ENSAYGPTRNPWNVDRVPGGSGGGSAAALAAFQAPLAIGSDTGGSIRQPA
....:* ***** :.****.******:** .. ***** .**:* **
Mflv_2712|M.gilvum_PYR-GCK AWTHLVGIKPQRGRISTSPLPEAFNGITVNGVLARTVSDAAMVLDAAAGN
Mvan_3824|M.vanbaalenii_PYR-1 AWTHLVGIKPQRGRISTWPLTEAFNGITVHGVLARTVSDAALLLDAASGN
TH_4482|M.thermoresistible__bu AWTHLVGIKPQRGRISTWPLPEAFNGITVHGVLARTVTDAALVLDAASGN
MMAR_3673|M.marinum_M AWTHLVGIKPQRGRISTWPWPETFNGITVNGVLARTVADAALVLDAASGN
MUL_3616|M.ulcerans_Agy99 AWTHLVGIKPQRGRISTWPWPETFNGITVNGVLARTVADAALVLDAASGN
Mb2384|M.bovis_AF2122/97 AWTHLVGIKPQRGRISTWPLPEAFNGVTVNGVLARTVEDAALVLDAASGN
Rv2363|M.tuberculosis_H37Rv AWTHLVGIKPQRGRISTWPLPEAFNGVTVNGVLARTVEDAALVLDAASGN
MAV_2032|M.avium_104 AWTHLVGIKPQRGRISTWPLPEAFNGITVNGVLARTVADAALVLDAASGN
MSMEG_4492|M.smegmatis_MC2_155 AWTHLVGIKPQRGRISTWPQPEAFNGVTVHGVLARTVVDAALVLDAASGN
MAB_1674c|M.abscessus_ATCC_199 AWTHLIGIKPQRGRISTWPFAEAFNGITVHGPLARTVADAALLLDAASGN
MLBr_01702|M.leprae_Br4923 ALTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHAVIAGH
* * :*:** * :* ..: * **** *:*:: . :*:
Mflv_2712|M.gilvum_PYR-GCK AAGDLHTPAPVTVSDFVSRAPGPLRVAVSTRFPFTGFR--ATLHPEIRSA
Mvan_3824|M.vanbaalenii_PYR-1 APGELHTPPPVRVSDYVSRAPGPLRIAMSTKFPFTVFR--AGLHPEIRTA
TH_4482|M.thermoresistible__bu APGDLHQPPPVRVSDYVGQAPGPLRIAVSTKFPFTLFR--AKLHPEIRAA
MMAR_3673|M.marinum_M VEGDLHKPAPLTASDYVGIAPGPLKIAMSTRFPYTGFR--AKLHPEILAA
MUL_3616|M.ulcerans_Agy99 AEGDLHKPAPLTASDYVGIAPGPLKIAMSTRFPYTGFR--AKLHPEILAS
Mb2384|M.bovis_AF2122/97 VEGDRHQPPPVTVSDFVGIAPGPLKIALSTHFPYTGFR--AKLHPEILAA
Rv2363|M.tuberculosis_H37Rv VEGDRHQPPPVTVSDFVGIAPGPLKIALSTHFPYTGFR--AKLHPEILAA
MAV_2032|M.avium_104 VDGDRHKPPPITASDYVGKAPGPLNIALSTRFPYTGFR--PKLHPEILAA
MSMEG_4492|M.smegmatis_MC2_155 ADGDLHKPAPIQASDYVGVAPGPLRIAMSTKIPFTGVR--AKIHPEILAA
MAB_1674c|M.abscessus_ATCC_199 VPGDLHKPPHVRVLDAVHEDPGQLRVALSLKVPFSGFP--AQLHPTIEAA
MLBr_01702|M.leprae_Br4923 DARDSTSVEAAVPDIVGAAKAGESGDLHGVRVGVVRQLRGEGYQSGVLAS
: .* . :. :. : ::
Mflv_2712|M.gilvum_PYR-GCK LDGVADQLSQLGHTVLNADP---DYTLGMSWNFLARSTSGIPEWMDRL--
Mvan_3824|M.vanbaalenii_PYR-1 LQNVADQLTQLGHTIVAADP---DYHLRMSWNFLSRSTSGIPEWMDRL--
TH_4482|M.thermoresistible__bu VQTVADQLEQLGHTVFPADP---EYTVGMSWSFLSRSTAGLLDWSRRL--
MMAR_3673|M.marinum_M TRRVGEQLQLLGHTVERGNP---DYGLRLSWDFLARSTAGLWEWAECL--
MUL_3616|M.ulcerans_Agy99 TRRVGEQLQLLGHTVERGNP---DYGLRLSWDFLARSTAGLWEWAECL--
Mb2384|M.bovis_AF2122/97 TQRVGDQLELLGHTVVKGNP---DYGLRLSWNFLARSTAGLWEWAERL--
Rv2363|M.tuberculosis_H37Rv TQRVGDQLELLGHTVVKGNP---DYGLRLSWNFLARSTAGLWEWAERL--
MAV_2032|M.avium_104 TRAVGKQLELLGHTVVPGNP---DYGMRLSWDFLARSTAGLRDWEERL--
MSMEG_4492|M.smegmatis_MC2_155 LQTIADHLGELGHTVKAKDP---NYSLRMSWDFLSRSTAGLLDWDDRLDT
MAB_1674c|M.abscessus_ATCC_199 TRYVAHQLRSLGHMVSEGDP---DYGVGLGLNFLPRATAGLLPWRDRL--
MLBr_01702|M.leprae_Br4923 FQAAVEQLIALGATVSEVDCPHFDYALAAYYLILPSEVSSNLARFDAMRY
.:* ** : : :* : :*. .:. :
Mflv_2712|M.gilvum_PYR-GCK -----GHRVTWDSRTRSNARMGRLLSQHTLRKARAREAAAQQRIGWIFNL
Mvan_3824|M.vanbaalenii_PYR-1 -----GRGVNWDSRTRSNARMGWLLSQNVLRKARAGEAASQQRIGWIFNL
TH_4482|M.thermoresistible__bu -----GDNVLWDKRTVANMRLGRLLSESALRKARAREAAAQRRMSWIFNI
MMAR_3673|M.marinum_M -----GDGVTLDPRTVTNMRAGHALSQAILRNARHHEAAAQRRVGSIFDI
MUL_3616|M.ulcerans_Agy99 -----GNGVTLDPRTVTNMRAGHALSQAILRNARHHEAAAQRRVGSIFDI
Mb2384|M.bovis_AF2122/97 -----GDEVTLDRRTVSNLRMGHVLSQAILRSARRHEAADQRRVGSIFDI
Rv2363|M.tuberculosis_H37Rv -----GDEVTLDRRTVSNLRMGHVLSQAILRSARRHEAADQRRVGSIFDI
MAV_2032|M.avium_104 -----GDGVVLDPRTVANLRLGHLLGQAILRSARRHEAADQRRVGSIFDI
MSMEG_4492|M.smegmatis_MC2_155 ----AGHGVVYDHRTVANMRIGRLLSQNVLRKARAHEAAVQRRMSWIFNL
MAB_1674c|M.abscessus_ATCC_199 -----DAGAHWDPRTVTNMRTGRRLAGWALRRARAAEPRLQARVGKIFGS
MLBr_01702|M.leprae_Br4923 GLRVGDDGSRSAEEVIALTRAAGFGPEVKRRIMIGAYALSAGYYDSYYSQ
. .. : * . * . . :.
Mflv_2712|M.gilvum_PYR-GCK ADVVVAPTTAQPPPEAHAFDGLGGLATDRMMIRACP-------------V
Mvan_3824|M.vanbaalenii_PYR-1 ADVVLAPTTALPPPEVRAFDDLGGLATDRMMIKACP-------------V
TH_4482|M.thermoresistible__bu ADVVLAPTTALPPPEVHAFDQLGGLATDRLMIKACP-------------V
MMAR_3673|M.marinum_M VDVVLAPTTAQPPPLARAFDDMGGLATDRAIIAACP-------------L
MUL_3616|M.ulcerans_Agy99 VDVVLAPTTAQPPPLARAFDDMGGLATDRAIIAACP-------------L
Mb2384|M.bovis_AF2122/97 VDVVLAPTTAQPPPMARAFDRLGSFGTDRAIIAACP-------------S
Rv2363|M.tuberculosis_H37Rv VDVVLAPTTAQPPPMARAFDRLGSFGTDRAIIAACP-------------S
MAV_2032|M.avium_104 VDVVLAPTTAQPPPLARAFDRLSGFGTDRAMIAACP-------------L
MSMEG_4492|M.smegmatis_MC2_155 VDVIIAPTTAQPPPPVHECDRLGWLATERKAIAACP-------------L
MAB_1674c|M.abscessus_ATCC_199 FNVVLAPTTAQPPTGIYDFDDVGSLATDRGQVGACP-------------M
MLBr_01702|M.leprae_Br4923 AQKVRTLIARDLDKAYRSVDVLVSPATPTTAFSLGEKADDPLAMYLFDLC
: : : : * : .* .
Mflv_2712|M.gilvum_PYR-GCK TWPWNLLGWPSINVPAGFTSDG-LPIGVQLMGPADSEPLLISLAAALEAI
Mvan_3824|M.vanbaalenii_PYR-1 TWPWNLLGWPSINVPAGFTSDG-LPIGVQLMGPANSEPLLVSLAAALEAI
TH_4482|M.thermoresistible__bu TWPWNLLGWPSINVPAGFTSDG-LPIGVQLMGPANSEPLLVSLAAALEGI
MMAR_3673|M.marinum_M TWPWNLLGWPSINVPAGFTVDG-LPIGVQLMGPAESEGLLISLAAELEAV
MUL_3616|M.ulcerans_Agy99 TWPWNLLGWPSINVPAGFTVDG-LPIGVQLMGPAESEGLLISLAAELEAV
Mb2384|M.bovis_AF2122/97 TWPWNLLGWPSINVPAGFTSDG-LPIGVQLMGPANSEGMLISLAAELEAV
Rv2363|M.tuberculosis_H37Rv TWPWNLLGWPSINVPAGFTSDG-LPIGVQLMGPANSEGMLISLAAELEAV
MAV_2032|M.avium_104 TFPWNVLGWPSINVPAGFTSDG-LPIGVQLMGPANSEGMLISLAAELEAV
MSMEG_4492|M.smegmatis_MC2_155 TWAWNLLGWPSINVPAGFTSDG-LPIGVQLMGPAESEPLLISLAAELEAI
MAB_1674c|M.abscessus_ATCC_199 TWPWNVLGWPSINVPAGFTADG-LPIGVQLMGPANSEPLLISLAAQLESI
MLBr_01702|M.leprae_Br4923 TLPLNLAGHCGMSVPSGLSSDDDLPVGLQIMAPALADDRLYRVGAAYEAA
* . *: * .:.**:*:: *. **:*:*:*.** :: * :.* *.
Mflv_2712|M.gilvum_PYR-GCK NGWATLQPENWWAPQ------------------PPTEPDL----TAPDSV
Mvan_3824|M.vanbaalenii_PYR-1 NGWAVHQPEDWWTPQ------------------PPPEPDLGSNVAAPASV
TH_4482|M.thermoresistible__bu NGWAARQPRPWWLT-------------------PPAAPXX----XASAAA
MMAR_3673|M.marinum_M SGWATKQPKVWWNTG-------------------TSTPPIHSVRPPRR--
MUL_3616|M.ulcerans_Agy99 SGWATKQPKVWWNTG-------------------TSTPPIHSVRPPRR--
Mb2384|M.bovis_AF2122/97 SGWATKQPQVWWTS------------------------------------
Rv2363|M.tuberculosis_H37Rv SGWATKQPQVWWTS------------------------------------
MAV_2032|M.avium_104 SGWASKQPQPWWDQS-------------------ADTPPAPGQPAPTG--
MSMEG_4492|M.smegmatis_MC2_155 TGWAAREPEVWWNTPEGRGLPPVSEITQRVPAPEDEKPAVETTDKPSEPV
MAB_1674c|M.abscessus_ATCC_199 NNWASEIPDPWW--------------------------------------
MLBr_01702|M.leprae_Br4923 RGPLPSAI------------------------------------------
.
Mflv_2712|M.gilvum_PYR-GCK TGQVA---------
Mvan_3824|M.vanbaalenii_PYR-1 EGQVA---------
TH_4482|M.thermoresistible__bu S-------------
MMAR_3673|M.marinum_M --------------
MUL_3616|M.ulcerans_Agy99 --------------
Mb2384|M.bovis_AF2122/97 --------------
Rv2363|M.tuberculosis_H37Rv --------------
MAV_2032|M.avium_104 --------------
MSMEG_4492|M.smegmatis_MC2_155 DDVSQAVAAVNALR
MAB_1674c|M.abscessus_ATCC_199 --------------
MLBr_01702|M.leprae_Br4923 --------------