For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AKSARRLKSADFPQLPQAPDDYPTFPDTSSWPVVFPELPPSPDGGPRRPPQHISKTAAPRIPADRLPNHV AIVMDGNGRWATERGLRRTEGHKMGEAVVIDIACGAVELGIKWLSLYAFSTENWKRSAEEVRFLMGFNRD VVRRRRDVLNQMGVRIRWVGSRPRLWRSVINELAVAEAMTNNNDVITINYCVNYGGRTEIAEATRDIARE VAAGRLNPERITESTIARHMQRPDIPDVDLFLRTSGEQRSSNFMLWQSAYAEYIFQDKLWPDYDRRDLWA ACEEYASRNRRFGSA*
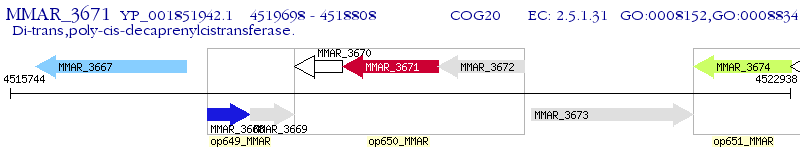
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3671 | - | - | 100% (296) | long (C50) chain Z-isoprenyl diphosphate synthase |
| M. marinum M | MMAR_2589 | - | 3e-42 | 40.43% (235) | Z-decaprenyl diphosphate synthase |
| M. marinum M | MMAR_4380 | uppS | 4e-33 | 35.17% (236) | short (C15) chain Z-isoprenyl diphosphate synthase, UppS |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2382c | - | 1e-159 | 89.53% (296) | long (C50) chain Z-isoprenyl diphosphate synthase (Z-decaprenyl |
| M. gilvum PYR-GCK | Mflv_2714 | - | 1e-108 | 76.09% (230) | undecaprenyl diphosphate synthase |
| M. tuberculosis H37Rv | Rv2361c | - | 1e-159 | 89.53% (296) | long (C50) chain Z-isoprenyl diphosphate synthase (Z-decaprenyl |
| M. leprae Br4923 | MLBr_00634 | - | 1e-155 | 86.49% (296) | undecaprenyl pyrophosphate synthase |
| M. abscessus ATCC 19977 | MAB_1676 | - | 1e-108 | 63.19% (288) | undecaprenyl pyrophosphate synthetase |
| M. avium 104 | MAV_2034 | uppS | 1e-153 | 85.14% (296) | undecaprenyl diphosphate synthase |
| M. smegmatis MC2 155 | MSMEG_4490 | uppS | 1e-135 | 76.06% (284) | undecaprenyl diphosphate synthase |
| M. thermoresistible (build 8) | TH_0839 | - | 1e-137 | 77.46% (284) | LONG (C50) CHAIN Z-ISOPRENYL DIPHOSPHATE SYNTHASE (Z-DECAPRENYL |
| M. ulcerans Agy99 | MUL_3614 | - | 1e-176 | 100.00% (296) | long (C50) chain Z-isoprenyl diphosphate synthase |
| M. vanbaalenii PYR-1 | Mvan_3822 | - | 1e-110 | 77.83% (230) | undecaprenyl diphosphate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4490|M.smegmatis_MC2_155 ---MATT--RGKKTYPQLPPAPDDY-PTFPDKSTWPVVFPEIPAGTNGRF
Mvan_3822|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2714|M.gilvum_PYR-GCK --------------------------------------------------
TH_0839|M.thermoresistible__bu -------------VYPQLPAPPADY-PTFPDKSTWPVVFPELPPNTNGRF
MMAR_3671|M.marinum_M MAKSARR--LKSADFPQLPQAPDDY-PTFPDTSSWPVVFPELPPSPDGGP
MUL_3614|M.ulcerans_Agy99 MAKSARR--LKSADFPQLPQAPDDY-PTFPDTSSWPVVFPELPPSPDGGP
Mb2382c|M.bovis_AF2122/97 MARDARK--RTSSNFPQLPPAPDDY-PTFPDTSTWPVVFPELPAAPYGGP
Rv2361c|M.tuberculosis_H37Rv MARDARK--RTSSNFPQLPPAPDDY-PTFPDTSTWPVVFPELPAAPYGGP
MLBr_00634|M.leprae_Br4923 MVRNERT--LKSTDFPQLPPAPDDY-PTFPDKSTWPVVFPMLPPSPDGGP
MAV_2034|M.avium_104 MVRAERA--PKSTGFPQLPPAPDDY-PVFPDKSTWPVVFPELPPSPGGGP
MAB_1676|M.abscessus_ATCC_1997 MVKSSQPDPQQDKYYTDWADVPQGYNPTFPDKTVFPAVFPKLLPGVGPGG
MSMEG_4490|M.smegmatis_MC2_155 ARP---PQHTSKAAAPKIPADQVPNHVAVVMDGNGRWATQRGLGRTEGHK
Mvan_3822|M.vanbaalenii_PYR-1 ----------------------MPNHVAVVMDGNGRWATQRGLGRTEGHK
Mflv_2714|M.gilvum_PYR-GCK ----------------------MPQHVAVVMDGNGRWATQRGLGRTEGHK
TH_0839|M.thermoresistible__bu ARP---PQHPSRAVAPRIPADQVPNHVAVVMDGNGRWATQRGLHRTEGHK
MMAR_3671|M.marinum_M RRP---PQHISKTAAPRIPADRLPNHVAIVMDGNGRWATERGLRRTEGHK
MUL_3614|M.ulcerans_Agy99 RRP---PQHISKTAAPRIPADRLPNHVAIVMDGNGRWATERGLRRTEGHK
Mb2382c|M.bovis_AF2122/97 CRP---PQHTSKAAAPRIPADRLPNHVAIVMDGNGRWATQRGLARTEGHK
Rv2361c|M.tuberculosis_H37Rv CRP---PQHTSKAAAPRIPADRLPNHVAIVMDGNGRWATQRGLARTEGHK
MLBr_00634|M.leprae_Br4923 RRP---PQHTSKAVAPRLSAAQLPTHVAIVMDGNGRWANQRGLHRTEGHK
MAV_2034|M.avium_104 SRP---PQHISKAAAPRIPADRLPNHVAIVMDGNGRWATERGMSRTDGHK
MAB_1676|M.abscessus_ATCC_1997 TRPHPAPKHPSGAVPPAIPKELIPRHVAVVMDGNGRWARARGLPRTEGHK
:* ***:********* **: **:***
MSMEG_4490|M.smegmatis_MC2_155 MGEAVLIDITCGAIEIGIKHLTVYAFSTENWKRSTEEVRFLMGFNREVVR
Mvan_3822|M.vanbaalenii_PYR-1 MGEAVLIDITCGAIEIGIKHLTVYAFSTENWKRSTEEVRFLMGFNREVVR
Mflv_2714|M.gilvum_PYR-GCK MGEAVLIDITCGAIEIGIKHLTVYAFSTENWKRSTEEVRFLMGFNREVVR
TH_0839|M.thermoresistible__bu MGEAVLIDITCGAIEIGIKHLTVYAFSTENWKRSPEEVRFLMGFNREVVR
MMAR_3671|M.marinum_M MGEAVVIDIACGAVELGIKWLSLYAFSTENWKRSAEEVRFLMGFNRDVVR
MUL_3614|M.ulcerans_Agy99 MGEAVVIDIACGAVELGIKWLSLYAFSTENWKRSAEEVRFLMGFNRDVVR
Mb2382c|M.bovis_AF2122/97 MGEAVVIDIACGAIELGIKWLSLYAFSTENWKRSPEEVRFLMGFNRDVVR
Rv2361c|M.tuberculosis_H37Rv MGEAVVIDIACGAIELGIKWLSLYAFSTENWKRSPEEVRFLMGFNRDVVR
MLBr_00634|M.leprae_Br4923 MGEAVVIDVACGAIELGIKWLSLYAFSTENWKRSVEEVRFLMGFNRDVVR
MAV_2034|M.avium_104 AGEAVVIDIVCGAIEIGIKWLSLYAFSTENWKRSPEEVRFLMGFNRDVVR
MAB_1676|M.abscessus_ATCC_1997 MGEATMMDMVCGAIEMGIPWLTTYAFSTENWRRPADEVRFLMGFNRDVVK
***.::*:.***:*:** *: ********:*. :**********:**:
MSMEG_4490|M.smegmatis_MC2_155 RRRENLNDMGVRMRWVGSRPRMWRSVIKEFDIAEQMTVDNDVITINYCVN
Mvan_3822|M.vanbaalenii_PYR-1 RRRENLNDMGVRMRWVGSRPRMWRSVIKEFDIAEQMTVDNDVITINYCVN
Mflv_2714|M.gilvum_PYR-GCK RRRENLNDMGVRMRWVGSRPRMWSSVIKEFDIAEQMTVDNDVITINYCVN
TH_0839|M.thermoresistible__bu RRRENLNDMGVKMRWVGSRPRMWRSVIKEFEIAENMTLDNDVITINYCVN
MMAR_3671|M.marinum_M RRRDVLNQMGVRIRWVGSRPRLWRSVINELAVAEAMTNNNDVITINYCVN
MUL_3614|M.ulcerans_Agy99 RRRDVLNQMGVRIRWVGSRPRLWRSVINELAVAEAMTNNNDVITINYCVN
Mb2382c|M.bovis_AF2122/97 RRRDTLKKLGVRIRWVGSRPRLWRSVINELAVAEEMTKSNDVITINYCVN
Rv2361c|M.tuberculosis_H37Rv RRRDTLKKLGVRIRWVGSRPRLWRSVINELAVAEEMTKSNDVITINYCVN
MLBr_00634|M.leprae_Br4923 RRRENLKEMGVRIRWVGSRPRLWRSVINELAIAEHMTAGNDVITINYCVN
MAV_2034|M.avium_104 MRRVNLNTIGVKIRWVGSRPRLWRSVINELAIAEEMTKRNDVITVNYCVN
MAB_1676|M.abscessus_ATCC_1997 RRRHDLNNMGVRFRWAGQKTRLWSSVYKELEITEELTKNNSTLTLTTCIN
** *: :**::**.*.:.*:* ** :*: ::* :* *..:*:. *:*
MSMEG_4490|M.smegmatis_MC2_155 YGGRTEIVEAARALAQEAVDGKINPARISEAMFAKHLHRADIPDVDLFIR
Mvan_3822|M.vanbaalenii_PYR-1 YGGRTEIVEAARELAQQAVDGKINPARITEATFAKHLHRADIPDVDLFIR
Mflv_2714|M.gilvum_PYR-GCK YGGRTEIVEAARELAQQAVDGKIKPNRITEAQFAKHLHRSDIPDVDLFIR
TH_0839|M.thermoresistible__bu YGGRAEITEAARSIAKEAVAGRINPDRITEKTVARHLHRPDIPDVDLFIR
MMAR_3671|M.marinum_M YGGRTEIAEATRDIAREVAAGRLNPERITESTIARHMQRPDIPDVDLFLR
MUL_3614|M.ulcerans_Agy99 YGGRTEIAEATRDIAREVAAGRLNPERITESTIARHMQRPDIPDVDLFLR
Mb2382c|M.bovis_AF2122/97 YGGRTEITEATREIAREVAAGRLNPERITESTIARHLQRPDIPDVDLFLR
Rv2361c|M.tuberculosis_H37Rv YGGRTEITEATREIAREVAAGRLNPERITESTIARHLQRPDIPDVDLFLR
MLBr_00634|M.leprae_Br4923 YGGRTEIAEATREIARLAAAGRLNPERITESTITRHLQRPDIPDVDLFVR
MAV_2034|M.avium_104 YGGRAEIAEATKEIARLAAAGRLNPERITESTVAHHLQRPDIPDVDLFLR
MAB_1676|M.abscessus_ATCC_1997 YGGRAEIAEATRRIARLVESGQLKPDRITEETIAKYLDEPDMPDVDLFLR
****:**.**:: :*: . *:::* **:* .::::...*:******:*
MSMEG_4490|M.smegmatis_MC2_155 TSGEQRASNFLLWQAAYAEYVFQDKLWPDYDRRDLWAACEEYVNRNRRFG
Mvan_3822|M.vanbaalenii_PYR-1 TSGEQRASNFLLWQAAYAEYVFQDKLWPDYDRRDLWAACEEYVNRNRRFG
Mflv_2714|M.gilvum_PYR-GCK TSGEQRASNFLLWQAAYAEYVFQDKLWPDYDRRDLWAACEEYVHRNRRFG
TH_0839|M.thermoresistible__bu TSGEQRASNFLLWQSAYAEFVFQDKLWPDYDRRDLWAACEEYVRRNRRFG
MMAR_3671|M.marinum_M TSGEQRSSNFMLWQSAYAEYIFQDKLWPDYDRRDLWAACEEYASRNRRFG
MUL_3614|M.ulcerans_Agy99 TSGEQRSSNFMLWQSAYAEYIFQDKLWPDYDRRDLWAACEEYASRNRRFG
Mb2382c|M.bovis_AF2122/97 TSGEQRSSNFMLWQAAYAEYIFQDKLWPDYDRRDLWAACEEYASRTRRFG
Rv2361c|M.tuberculosis_H37Rv TSGEQRSSNFMLWQAAYAEYIFQDKLWPDYDRRDLWAACEEYASRTRRFG
MLBr_00634|M.leprae_Br4923 TSGEQRSSNFMLWQAAYTEYIFQDKLWPDYDRRDLWAACEEYASRNRRFG
MAV_2034|M.avium_104 TSGEQRSSNFMLWQAAYAEYIFQDKLWPDYDRRDLWAACQEYASRNRRFG
MAB_1676|M.abscessus_ATCC_1997 PSGEFRSSNFMMWQSAYAEFVFQEKMYPDFDRRDLWAACLEYAQRDRRFG
.*** *:***::**:**:*::**:*::**:********* **. * ****
MSMEG_4490|M.smegmatis_MC2_155 RA-
Mvan_3822|M.vanbaalenii_PYR-1 RAE
Mflv_2714|M.gilvum_PYR-GCK RA-
TH_0839|M.thermoresistible__bu RA-
MMAR_3671|M.marinum_M SA-
MUL_3614|M.ulcerans_Agy99 SA-
Mb2382c|M.bovis_AF2122/97 SA-
Rv2361c|M.tuberculosis_H37Rv SA-
MLBr_00634|M.leprae_Br4923 SA-
MAV_2034|M.avium_104 SA-
MAB_1676|M.abscessus_ATCC_1997 SA-
*