For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LATENSTPDAGFQAYRCGSQAIHIPLWGNIFGNAIGIHMVFWSRLKPRRRAVSPVRALLPLSRDLQTGPN SQYRRSYLDPERPGPASTDPSPARLAGESAASPRRLPIMMVRKFIDDLAASLRCGSSARPVLFGFMMISN LAGVALVLVLVIVAIPTPDMFAPEVLWITEGVLPVYVVMAMTFGWVWLIKRVLAGLRWENDNQPLTAKDQ RNAFLAPWRLALVPVVSWTIGSALFAVLYGRVSYEYVPKIILGTSLSGIALSATCYFFAELMLRPAATIA LGGGQPTRRLAPSVMGRATTSWVLGSGVPLVGIILAVAFAMSQQHLTRNDLGLVLLLLAPAALVSGVLCN MIAAWLTATSVRDLHSALKRVESGDLDCQLDVFDGTELGELQRGFNAMVEGLRQRERIRDLFGRHVGRDV AAAAEDQSFELGGEERCVAVLFVDIIGSTRLVTARPPAEVVRLFNQMFAVIIDEVSRRNGLINKFQGDGT LAIFGAPIRALSPETDALGAARAIAHRLANEVPECQAAIGVAAGAVLAGNVGAFERFEYTVIGEPVNIAA RLCELAKSDPHRVLSSAETVQASAPDESQHWTLGGHTVLRGLDRLTQLASPNDCPTGPRDTHVVESLPAQ IRTSGFPSNMVR
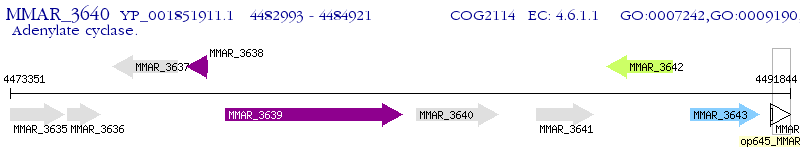
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3640 | - | - | 100% (642) | adenylate cyclase |
| M. marinum M | MMAR_4078 | - | e-142 | 51.56% (481) | adenylate cyclase |
| M. marinum M | MMAR_4120 | - | e-139 | 52.18% (481) | adenylate cyclase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1352c | - | 1e-145 | 53.22% (481) | adenylate cyclase |
| M. gilvum PYR-GCK | Mflv_2336 | - | 1e-143 | 52.17% (483) | putative adenylate/guanylate cyclase |
| M. tuberculosis H37Rv | Rv1318c | - | 1e-145 | 53.22% (481) | adenylate cyclase |
| M. leprae Br4923 | MLBr_00201 | - | 8e-88 | 39.29% (504) | hypothetical protein MLBr_00201 |
| M. abscessus ATCC 19977 | MAB_1459c | - | 1e-138 | 51.24% (482) | hypothetical protein MAB_1459c |
| M. avium 104 | MAV_1540 | - | 1e-149 | 55.28% (483) | HAMP domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_4924 | - | 1e-145 | 53.85% (481) | adenylate cyclase, family protein 3 |
| M. thermoresistible (build 8) | TH_3474 | - | 1e-147 | 54.05% (481) | PUTATIVE putative adenylate/guanylate cyclase |
| M. ulcerans Agy99 | MUL_1472 | - | 0.0 | 98.50% (533) | adenylate cyclase |
| M. vanbaalenii PYR-1 | Mvan_4309 | - | 1e-142 | 51.77% (481) | putative adenylate/guanylate cyclase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3640|M.marinum_M MATENSTPDAGFQAYRCGSQAIHIPLWGNIFGNAIGIHMVFWSRLKPRRR
MUL_1472|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2336|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4309|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3474|M.thermoresistible__bu --------------------------------------------------
Mb1352c|M.bovis_AF2122/97 --------------------------------------------------
Rv1318c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_1540|M.avium_104 --------------------------------------------------
MSMEG_4924|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1459c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00201|M.leprae_Br4923 --------------------------------------------------
MMAR_3640|M.marinum_M AVSPVRALLPLSRDLQTGPNSQYRRSYLDPERPGPASTDPSPARLAGESA
MUL_1472|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2336|M.gilvum_PYR-GCK -------------------------MN--ATKSVPYRLGRVLERVTRQSG
Mvan_4309|M.vanbaalenii_PYR-1 -------------------------MN--ATKSVPYRLGRVLERVTRQSG
TH_3474|M.thermoresistible__bu -------------------------MT--PNNRLARRLGHLLETLTRQSG
Mb1352c|M.bovis_AF2122/97 -------------------------MS--AKKSTAQRLGRVLETVTRQSG
Rv1318c|M.tuberculosis_H37Rv -------------------------MS--AKKSTAQRLGRVLETVTRQSG
MAV_1540|M.avium_104 -------------------------MS--ATQSTAQRIGRVLEKITRQSG
MSMEG_4924|M.smegmatis_MC2_155 -------------------------MN--ADNSLARQLGRVLETVTRQNS
MAB_1459c|M.abscessus_ATCC_199 -------------------------MANESIRAVSRRVGRALGAITGQTG
MLBr_00201|M.leprae_Br4923 --------------------------------------------MATQAA
MMAR_3640|M.marinum_M ASPRRLPIMMVRKFIDDLAASLRCGSSARPVLFGFMMISNLAGVALVLVL
MUL_1472|M.ulcerans_Agy99 ---------MVRKFTDDLAAPLRCGSSARPVLFGFMMISNLAGVVLVLVL
Mflv_2336|M.gilvum_PYR-GCK RL-DTP-GYGSWLLGRPAESQSRRRVRIQIILTFFILFTNIMGIAVSLLL
Mvan_4309|M.vanbaalenii_PYR-1 RL-ETP-DYGSWLLGKPEESQARRRIRIQVILTFLLLFTNILGIAVSLLL
TH_3474|M.thermoresistible__bu RL-SGANEYGSWLLGRVSESQRRRRLRIQTILTTFIVGANVVGIAVAVTL
Mb1352c|M.bovis_AF2122/97 RL-PETPAYGSWLLGRVSESQRRRRVRIQVMLTALVVTANLLGIGVALLL
Rv1318c|M.tuberculosis_H37Rv RL-PETPAYGSWLLGRVSESQRRRRVRIQVMLTALVVTANLLGIGVALLL
MAV_1540|M.avium_104 RL-PETPAYGSLLLGRVTESQHRRRIRIQIIMTVMVLGANLIGIGAALIL
MSMEG_4924|M.smegmatis_MC2_155 RLPSGTPEYGSWILGRVTESQSRRRIRIQLILTTFVVIANLLGIAVSFLV
MAB_1459c|M.abscessus_ATCC_199 NINTGN-GYGSWLLGTADESTFIQRIRIQLILTVFVVAANLIGIGVVILL
MLBr_00201|M.leprae_Br4923 GLPGRIGAFVRWGIRTPWPLFSLSMLQSDIIGALFVLGFLRYGLPPQDRV
: : ::: *: :
MMAR_3640|M.marinum_M VIVAIPTPDMFAPEVLWITEGVLPVYVVMAMTFGWVWLIKRVLAGLRWEN
MUL_1472|M.ulcerans_Agy99 VIVAIPTPDMFAPEVLWITEGVLPVYVVMAMTFGWVWLIKRVLAGLRWEN
Mflv_2336|M.gilvum_PYR-GCK NTIAIPVPSVFSDAPAWLTFGVTPAYMLVALFFGTAWITSRTVRSLRWAI
Mvan_4309|M.vanbaalenii_PYR-1 NTVAIPVPSVFSDAPAWLTFGVVPAYMVVALVLGTAWITSRTVGSLRWAI
TH_3474|M.thermoresistible__bu VSFVFPVPSIFSDAPAWLTFGVSPALVGVALVVGTTWITRGIVRNLRWAI
Mb1352c|M.bovis_AF2122/97 VTIAIPEPSIVRDTPRWLTFGVVPGYVLLALALGSYALTRQTVQALRWAI
Rv1318c|M.tuberculosis_H37Rv VTIAIPEPSIVRDTPRWLTFGVVPGYVLLALALGSYALTRQTVQALRWAI
MAV_1540|M.avium_104 VIVAIPAPSVFNDAPAWITFGVGPAYVASALAVGTYWITRRTVLALRWAI
MSMEG_4924|M.smegmatis_MC2_155 LTVLFPSPSVFDARVAPYTYIVAPAYVVGALVVGVIWATNRVIGNVRWAL
MAB_1459c|M.abscessus_ATCC_199 VTVAFPTPNVF-DAPAWVNFVIVPVYIALAVLIGAVWGTRRIFKAVRWAL
MLBr_00201|M.leprae_Br4923 ELQDLPIRNLAISTLSVVVAFSTGFAVNLKLLMPVFRWQRR---DNRLAE
:* .: : : . *
MMAR_3640|M.marinum_M DNQPLTAKDQRNAFLAPWRLALVPVVSWTIGSALFAVLYGRVSYEYVPKI
MUL_1472|M.ulcerans_Agy99 DNQPLTAKDQRNAFLAPWRLALVPVVSWTIGSALFAVLYGRVSYEYVPKI
Mflv_2336|M.gilvum_PYR-GCK EERQPTSTDQRNVFLAPARVAQKLLLLWGVGAVLLTVLYGLQDTAFIPRF
Mvan_4309|M.vanbaalenii_PYR-1 EERSPTRTDQRNVFVAPSRVAQMLLLFWGFGAALLTLLYGLQDSAFIPRF
TH_3474|M.thermoresistible__bu EERPPSRTDQRNTFLAPWRVARAHLVLWGAGTAVLTTLYGLVDPAFIPRY
Mb1352c|M.bovis_AF2122/97 EGRKPTREEERRTFLAPWRVAVGHLMFWGVGTALLTTLYGLINNAFIPRF
Rv1318c|M.tuberculosis_H37Rv EGRKPTREEERRTFLAPWRVAVGHLMFWGVGTALLTTLYGLINNAFIPRF
MAV_1540|M.avium_104 EERKPTPTDERNTFLAPWRIALVDLVLWGAGTLLLTVLYGLVDTMFIPRF
MSMEG_4924|M.smegmatis_MC2_155 EERPPTPQDQRNTFFAPFRLTRVLLILWAAGTALLTTLYGMVDTNFIPKV
MAB_1459c|M.abscessus_ATCC_199 DERPPTKADQRNAFAVPWRLTVIQSVLWAGGVALFTLLYGLYDPMYIPKI
MLBr_00201|M.leprae_Br4923 ADPAATELARSRALRMPFYHTIISLVTWGIGGAVFIIASWSAARHSAPIV
: . ..: * : : * * :: *
MMAR_3640|M.marinum_M ILGTSLSGIALSATCYFFAELMLRPAATIALGGGQPTRRLAPSVMGRATT
MUL_1472|M.ulcerans_Agy99 ILGTSLSGIALSATCYFFAELMLRPAATIALGGGQPTRRLAPSVMGRATT
Mflv_2336|M.gilvum_PYR-GCK LFAVGFPGIVVATACYMITEFALRPIAAQALEAGRPPRRLAHGVMGRTMT
Mvan_4309|M.vanbaalenii_PYR-1 LFAVGFPGIVVATACYMITEFALRPVAAQALEAGRPPRRLAHGVMGRTMT
TH_3474|M.thermoresistible__bu LFTVSFAGIVVATICYLFTEFALRPVAAQALEAGPPPRRLAHGIMGRTMT
Mb1352c|M.bovis_AF2122/97 LFAVSFCGVLVATATYLHTEFALRPFAAQALEAGPPPRRLAPGILGRTMV
Rv1318c|M.tuberculosis_H37Rv LFAVSFCGVLVATATYLHTEFALRPFAAQALEAGPPPRRLAPGILGRTMV
MAV_1540|M.avium_104 LFGVSFCGVLVATACYLLAEFALRPVAAQALEAGPPPRRLTAGIMGRTMT
MSMEG_4924|M.smegmatis_MC2_155 LLGISFPGIVVSVSCHLFTEFALRPVAAQALEAGPPPVRLAPGIMGRTMV
MAB_1459c|M.abscessus_ATCC_199 FFAVGFSGAVVCAANYLFTEFAMRPVAARALEAGYRRRRLASGVMVRTVL
MLBr_00201|M.leprae_Br4923 ALATALGATATAIIGYLQSERVLRPVAVAALRSGVPENVKTPGVTLRLML
: .: . . :: :* :** *. ** .* : .: *
MMAR_3640|M.marinum_M SWVLGSGVPLVGIILAVAFA-MSQQHLTRNDLGLVLLLLAPAALVSGVLC
MUL_1472|M.ulcerans_Agy99 SWVLGSGVPLVGIILAVAFA-MSQQHLTRNDLGLVLLLLAPAALVSGVLC
Mflv_2336|M.gilvum_PYR-GCK VWLLSSGVPVLGILLLAVFS-LSLKNLSATQFAVAVIFIAAFALVFGLTL
Mvan_4309|M.vanbaalenii_PYR-1 VWLLSSGVPVLGILLLAIFT-LSLRNLSATQFAVAVMIISVVALMFGLIL
TH_3474|M.thermoresistible__bu VWLLGSGVPLLGIMIAAAVS-LTQRHLSLVQFTIATMVLAAVALVFGFVL
Mb1352c|M.bovis_AF2122/97 VWLLGSGVPVVGIALMAMFE-MVLLNLTRMQFATGVLIISMVTLVFGFIL
Rv1318c|M.tuberculosis_H37Rv VWLLGSGVPVVGIALMAMFE-MVLLNLTRMQFATGVLIISMVTLVFGFIL
MAV_1540|M.avium_104 VWFLGSGVPVIGIALLALFE-IWLRNLTETEFAVGVLIVSTAALIFGCLL
MSMEG_4924|M.smegmatis_MC2_155 VWALGSGVPILGILLAGVFA-VTLENLTPTQFAYAVMGLAVFALVFGFIL
MAB_1459c|M.abscessus_ATCC_199 AWMLGSGVPAAGIVITGIGA-LVLHNLTVSQLAVAVIIIAGAIPSFGLIL
MLBr_00201|M.leprae_Br4923 TWVPSTGVPVLAIVLAVVADKIALLHAVPEQLFSPILLLALAVLVVGLVS
* .:*** .* : : : :: : :: *
MMAR_3640|M.marinum_M NMIAAWLTATSVRDLHSALKRVESGDLDCQLDVFDGTELGELQRGFNAMV
MUL_1472|M.ulcerans_Agy99 NMIAAWLTATSVRDLHSALKRVESGDLDCQLDVFDGTELGELQRGFNAMV
Mflv_2336|M.gilvum_PYR-GCK MWLLSWLIATPVRVVRSALKQVEDGDLDCNLVVFDGTELGELQRGFNSMV
Mvan_4309|M.vanbaalenii_PYR-1 MWLLSWLIATPVRVVRSALKHVEDGDLDCNVVVFDGTELGELQRGFNSMV
TH_3474|M.thermoresistible__bu MWLVSWLTATPVRVVREALKQVENGNLDCNVVVFDGTELGELQRGFNAMV
Mb1352c|M.bovis_AF2122/97 MWILAWLTATPVRVVRAALRRVERGELRTNLVVFDGTELGELQRGFNAMV
Rv1318c|M.tuberculosis_H37Rv MWILAWLTATPVRVVRAALRRVERGELRTNLVVFDGTELGELQRGFNAMV
MAV_1540|M.avium_104 MWILAWLTATPVRVVRAALKRVERGDLRGDLVVFDGTELGELQRGFNAMV
MSMEG_4924|M.smegmatis_MC2_155 MWILSWLTATPIRVVRAALNRVEQGDLDTNLVVFDGTELGQLQRGFNSMV
MAB_1459c|M.abscessus_ATCC_199 IWIASWLMATPVKEVQTAIKRVAQNDLDIDLVVYDGTELGELQAGFNTMV
MLBr_00201|M.leprae_Br4923 TWLVAMSIADPLRQLRWALSEVQRGNYNAHMQIYDASELGLLQAGFNDMV
: : * .:: :: *: .* .: .: ::*.:*** ** *** **
MMAR_3640|M.marinum_M EGLRQRERIRDLFGRHVGRDVAAAAEDQSFELGGEERCVAVLFVDIIGST
MUL_1472|M.ulcerans_Agy99 EGLRQRERIRDLFRRHVGRDGAAAAEDQSFELGGEERCVAVLFVDIIGST
Mflv_2336|M.gilvum_PYR-GCK HGLRERERVRDLFGRHVGREVALAAEKQQVELGGEERRAAVIFVDIIGST
Mvan_4309|M.vanbaalenii_PYR-1 HGLRERERVRDLFGRHVGREVARAAEKQKIELGGEERHAAVIFVDIIGST
TH_3474|M.thermoresistible__bu DGLRERERVRDLFGRHVGREVARAAEEQQLELGGEERHVAVIFVDIVGST
Mb1352c|M.bovis_AF2122/97 AGLRERERVRDLFGRHVGREVAAAAERERSKLGGEERHVAVVFIDIVGST
Rv1318c|M.tuberculosis_H37Rv AGLRERERVRDLFGRHVGREVAAAAERERSKLGGEERHVAVVFIDIVGST
MAV_1540|M.avium_104 DGLRERERVRDLFGRHVGREVALAAERERPKLGGEERHVAVVFIDIVGST
MSMEG_4924|M.smegmatis_MC2_155 HGLRERERVRDLFGRHVGREVAVLAEQQKFTLGGEERHAAVVFVDVIGST
MAB_1459c|M.abscessus_ATCC_199 EGLRERERVRDLFGKHVGREVAAAAEQQQPELGGEERHIAVLFADIVGST
MLBr_00201|M.leprae_Br4923 RELSERQRLRDLFGRYVGEDVARRALEHGTELGGQERDVAVLFVDLIGST
* :*:*:**** ::**.: * * . ***:** **:* *::***
MMAR_3640|M.marinum_M RLVTARPPAEVVRLFNQMFAVIIDEVSRRNGLINKFQGDGTLAIFGAPIR
MUL_1472|M.ulcerans_Agy99 RLVTARPPAEVVRLFNQMFAVIIDEVSRRNGLINKFQGDGTLAIFGAPIR
Mflv_2336|M.gilvum_PYR-GCK KLVTSRPAAEVVELLNRFFDVVVDEVDKHHGLVNKFEGDAVLAVFGAPVS
Mvan_4309|M.vanbaalenii_PYR-1 QLVTSRPAVEVVELLNRFFDVVVEEVDNHHGLVNKFEGDAVLAVFGAPVS
TH_3474|M.thermoresistible__bu QLVTTRPAAEVVSLLNRFFAVIVEEVDRHRGLVNKFEGDATLAIFGAPNR
Mb1352c|M.bovis_AF2122/97 QLVTSRPPADVVKLLNKFFAIVVDEVDRHHGLVNKFEGDASLTIFGAPNR
Rv1318c|M.tuberculosis_H37Rv QLVTSRPPADVVKLLNKFFAIVVDEVDRHHGLVNKFEGDASLTIFGAPNR
MAV_1540|M.avium_104 QLVTRRPPAEVVSVLNQFFGIVVEEVDRHCGLVNKFEGDATLAIFGAPNH
MSMEG_4924|M.smegmatis_MC2_155 QLVTSRPAVEVVELLNRFFAVIVDEVDAHRGLVNKFEGDAVLAVFGAPVA
MAB_1459c|M.abscessus_ATCC_199 RLAATRPAVEVVALLNRFFTVVVEEIDRYEGMVNKFEGDATLAVFGAPVR
MLBr_00201|M.leprae_Br4923 QLAETKPPTKVVHLLNEFFRVVVDTVGRHGGFVNKFQGDAALAIFGAPIE
:*. :*...** ::*.:* :::: :. *::***:**. *::****
MMAR_3640|M.marinum_M ALSPETDALGAARAIAHRLANEVPECQAAIGVAAGAVLAGNVGAFERFEY
MUL_1472|M.ulcerans_Agy99 ALLPETDALGAARAIAHRLANEVPECQAAIGVAAGAVLAGNVGAFERFEY
Mflv_2336|M.gilvum_PYR-GCK LDNAEAEALAAARAMSRRLAAEVPECTAGIGVASGQVVAGNVGAKERFEY
Mvan_4309|M.vanbaalenii_PYR-1 LDSAETEALSAARAMARRLRTEVPECPAGIGVAAGQVVAGNVGAKQRFEY
TH_3474|M.thermoresistible__bu LDNPEDAALAAARAIAARLRTEVPEVRAGIGVASGQVVAGNVGARERFEY
Mb1352c|M.bovis_AF2122/97 LPCPEDKALAAARAIADRLVNEMPECQAGIGVAAGQVIAGNVGARERFEY
Rv1318c|M.tuberculosis_H37Rv LPCPEDKALAAARAIADRLVNEMPECQAGIGVAAGQVIAGNVGARERFEY
MAV_1540|M.avium_104 LDCPEDAALTAARAIADRLANEMPQCRAGIGVAAGQVVAGNVGARERFEY
MSMEG_4924|M.smegmatis_MC2_155 LDNAEDEALGAARGIARRLREEVPELDAGIGVAAGQVVAGNVGAKQRFEY
MAB_1459c|M.abscessus_ATCC_199 QDRPESQALAAARAIARRLRTEVPECQAGIGVASGQAVAGNIGAHDRFEY
MLBr_00201|M.leprae_Br4923 HPDSAGAALSAARELHDELLPVLGSAEFGIGVSAGRAIAGHIGAQARFEY
. ** *** : .* : . .***::* .:**::** ****
MMAR_3640|M.marinum_M TVIGEPVNIAARLCELAKSDPHRVLSSAETVQASAPDESQHWTLGGHTVL
MUL_1472|M.ulcerans_Agy99 TVIGEPVNIAARLCELAKSDPHRVLSSAETVQASAPDESQHWILGRHTVL
Mflv_2336|M.gilvum_PYR-GCK TVIGEPVNEAARLCELSKKIDGHLVASADAVHNATETERRHWELGETVTL
Mvan_4309|M.vanbaalenii_PYR-1 TVIGEPVNEAARLCELAKNVPALLVASADAVGGATESERALWKLGETVTL
TH_3474|M.thermoresistible__bu TVIGEPVNEGARLCELAKTKPGLLVASADAVANAGEQERAHWVLGETVTL
Mb1352c|M.bovis_AF2122/97 TVIGEPVNEAARLCELAKSRPGKLLASAQAVDAASEEERARWSLGRHVKL
Rv1318c|M.tuberculosis_H37Rv TVIGEPVNEAARLCELAKSRPGKLLASAQAVDAASEEERARWSLGRHVKL
MAV_1540|M.avium_104 TVIGEPVNEAARLCELAKSHSGRLLATGDAIEGASEKERARWSLGDTVTL
MSMEG_4924|M.smegmatis_MC2_155 TVIGEPVNEAARLCEFAKSVPGHVVASSDTLANASESERARWSLGESVTL
MAB_1459c|M.abscessus_ATCC_199 TVIGDPVNEAARLCELSKTVPGHLVASLDTVLRAHHREALHWRSGDTVTL
MLBr_00201|M.leprae_Br4923 TVIGDPVNEAARLTELAKLEDGHVLASAIAVSGALDAEALCWNVGEIVEL
****:*** .*** *::* :::: :: : * * * . *
MMAR_3640|M.marinum_M RGLDRLTQLASPNDCPTGPRDTHVVESLPAQIRTSGFPSNMVR
MUL_1472|M.ulcerans_Agy99 RGLDRLTQLASPNDCPTGPRDTHVVESLPAQIRTSGFPSNMVR
Mflv_2336|M.gilvum_PYR-GCK RGHAEPTTLATPVDRD---------------------------
Mvan_4309|M.vanbaalenii_PYR-1 RGHDQPTRLATPAGEL---------------------------
TH_3474|M.thermoresistible__bu RGHNQPTRLALPA------------------------------
Mb1352c|M.bovis_AF2122/97 RGHDQPVRLAKPVGLTKPRR-----------------------
Rv1318c|M.tuberculosis_H37Rv RGHDQPVRLAKPVGLTKPRR-----------------------
MAV_1540|M.avium_104 RGHERPTRLASPAGAADRPAGTD--------------------
MSMEG_4924|M.smegmatis_MC2_155 RGLDEPTRLAVPV------------------------------
MAB_1459c|M.abscessus_ATCC_199 RGRTEPTELAVPV------------------------------
MLBr_00201|M.leprae_Br4923 RGRTAPTQLARPLNLAVPEQITSEVTG----------------
** . ** *