For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TETMVDTEVIARAVELASRAPSLHNSQPWRWVAASGAVDLFLDPHRMVTSADDSGREAVISCGALLDHFQ VAMAAAGWDSNVDLFPNPNNPDHLASIDFAAIDDIAQARRERAEAISRRRTDRRPFRAPGDWESFEPVLR SSVDTETVTLAVLAEDARPRLAEASRLTEALRRYDDAYHHELAWWTAPSRESEGIPQSALASGTAAREVA VNRSFPVDWRSESSSAGRHDHATIVVLSTPGDSRADALNCGAALSAVLLECTLAGLATCPVTHLTEMDAG RDTVRELTGGQAEPQVLVRVGIAPTGAAPELTPRRPLRDVLEIRR*
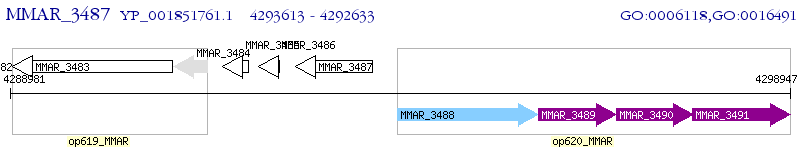
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3487 | - | - | 100% (326) | hypothetical protein MMAR_3487 |
| M. marinum M | MMAR_1522 | - | 5e-87 | 50.15% (327) | hypothetical protein MMAR_1522 |
| M. marinum M | MMAR_3475 | - | 2e-82 | 49.69% (324) | hypothetical protein MMAR_3475 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2058 | - | 1e-94 | 53.68% (326) | hypothetical protein Mb2058 |
| M. gilvum PYR-GCK | Mflv_3845 | - | 2e-89 | 52.74% (328) | hypothetical protein Mflv_3845 |
| M. tuberculosis H37Rv | Rv2032 | acg | 7e-96 | 53.99% (326) | hypothetical protein Rv2032 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3903 | - | 6e-86 | 49.09% (330) | hypothetical protein MAB_3903 |
| M. avium 104 | MAV_2505 | - | 1e-116 | 64.83% (327) | hypothetical protein MAV_2505 |
| M. smegmatis MC2 155 | MSMEG_3952 | - | 3e-99 | 56.29% (318) | hypothetical protein MSMEG_3952 |
| M. thermoresistible (build 8) | TH_1331 | - | 1e-101 | 57.23% (325) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_2417 | - | 6e-87 | 50.00% (330) | hypothetical protein MUL_2417 |
| M. vanbaalenii PYR-1 | Mvan_1383 | - | 1e-93 | 54.60% (326) | hypothetical protein Mvan_1383 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2058|M.bovis_AF2122/97 -MPDTMVTTDVIKSAVQLACRAPSLHNSQPWRWIAEDHTVA--------L
Rv2032|M.tuberculosis_H37Rv -MPDTMVTTDVIKSAVQLACRAPSLHNSQPWRWIAEDHTVA--------L
Mflv_3845|M.gilvum_PYR-GCK -MSATQVRIETIIDAVDVACRAPSLHNSQPWRWVATDHGVD--------L
Mvan_1383|M.vanbaalenii_PYR-1 -MRDVAVDIEVITEALALACRAPSLFNSQPWRWVSEGAVVS--------L
MAB_3903|M.abscessus_ATCC_1997 -MPKTMVHIDVIQTAIQLACRAPSLHNTQPWRWIVDLGASAGD----LHL
MMAR_3487|M.marinum_M -MTETMVDTEVIARAVELASRAPSLHNSQPWRWV----AASGA----VDL
MAV_2505|M.avium_104 MMMQMTADTEVITAAIELACHAPSLHNSQPWRWV----AGSTG----VDL
MSMEG_3952|M.smegmatis_MC2_155 ----------MITEAVGLACRAPSLHNSQPWRWV----DHGTS----VDL
TH_1331|M.thermoresistible__bu -----MPSVTILERALELACRAPSLHNSQPWRWIVDRTDDDTE----VQL
MUL_2417|M.ulcerans_Agy99 ------MDMEVLKTAVLLACRAPSVHNSQPWRWVADRDRDSGSEVVAVHL
: *: :*.:***:.*:*****: *
Mb2058|M.bovis_AF2122/97 FLDKDRVLYATDHSGREALLGCGAVLDHFRVAMAAAGTTANVERFPNPND
Rv2032|M.tuberculosis_H37Rv FLDKDRVLYATDHSGREALLGCGAVLDHFRVAMAAAGTTANVERFPNPND
Mflv_3845|M.gilvum_PYR-GCK YADPDRMIRAADSTGREALISCGAVLDHFRVAMAAAGWSITVERLPEPSD
Mvan_1383|M.vanbaalenii_PYR-1 FLDDARAPRYTDTTGRELVISCGIVLDHFRVAMAAAGWKCNVDRFPNPNN
MAB_3903|M.abscessus_ATCC_1997 YLDRTRCVTRTDGAGREAIISCGAALDHFRVAMASAGWMTRIERIPDPDN
MMAR_3487|M.marinum_M FLDPHRMVTSADDSGREAVISCGALLDHFQVAMAAAGWDSNVDLFPNPNN
MAV_2505|M.avium_104 FVDPRRTVKSADKSGREAIISCGAALDHFRVAMAAAGWSSNVAQFPNPNN
MSMEG_3952|M.smegmatis_MC2_155 FLDHHRIVRSSDSSGREAHISCGAALDHLRVAMAAAGWVIHVDRFPNPNN
TH_1331|M.thermoresistible__bu FADHRRIGRATDRSGRELVISCGAALDHLRVAMAAAGWQAGVERFPNPNN
MUL_2417|M.ulcerans_Agy99 FADRGRTVPATDHSGRETLISCGAVLDHLGVAMTAAGWQPSITRFPTPDD
: * * :* :*** :.** ***: ***::** : :* *.:
Mb2058|M.bovis_AF2122/97 PLHLASIDFSPADFVTEGHRLRADAILLRRTDRLPFAEPPDWDLVESQLR
Rv2032|M.tuberculosis_H37Rv PLHLASIDFSPADFVTEGHRLRADAILLRRTDRLPFAEPPDWDLVESQLR
Mflv_3845|M.gilvum_PYR-GCK PRHIASIRFAAATEVTAEQKRRADAILVRRTDRLPLSAPRDWDVPGETGA
Mvan_1383|M.vanbaalenii_PYR-1 RDHLASIDFSAMDFVTDAHRQRAEAIARRRTDRLPFAAPPDWESFIAQLS
MAB_3903|M.abscessus_ATCC_1997 PDHLAAITFSPRTVVVVGDRRRADAILQRRTDRLPFETPPNWSQFARMLN
MMAR_3487|M.marinum_M PDHLASIDFAAIDDIAQARRERAEAISRRRTDRRPFRAPGDWESFEPVLR
MAV_2505|M.avium_104 LDHLASIDVVPTEYVARARRDLADAILRRRTNRLPFRAPKHWSALEPVLR
MSMEG_3952|M.smegmatis_MC2_155 REHLANIQFSPVAHVTDAMRDRASAITRRRTDRLPLHPPAHWDTVAPLVN
TH_1331|M.thermoresistible__bu PDHLATVRFTALPAVTPAHRERAEAISARRTDRLPFDPPTDWSTVETVLR
MUL_2417|M.ulcerans_Agy99 PNHLATVDFRPIEEVTETELKRAESILQRRTDRLAFDRRRYWDLFEGVLR
*:* : . . :. *.:* ***:* .: *.
Mb2058|M.bovis_AF2122/97 TTVTAD-TVRIDVIADDMRPELAAASKLTESLRLYDSSYHAELFWWTGAF
Rv2032|M.tuberculosis_H37Rv TTVTAD-TVRIDVIADDMRPELAAASKLTESLRLYDSSYHAELFWWTGAF
Mflv_3845|M.gilvum_PYR-GCK AGAEPDNPVRVHTVPGHKRDMLVEASQLAEVLRLYDSDYHSELFWWTADF
Mvan_1383|M.vanbaalenii_PYR-1 SGTAQG-SVSLHTIADGSRPELVKASELTRSARRYDSKYHAELIGWTADV
MAB_3903|M.abscessus_ATCC_1997 TVIADR-AVLLDVIDDVMRPKLADASALSDALRIYDSAYQTELDWWTAPF
MMAR_3487|M.marinum_M SSVDTE-TVTLAVLAEDARPRLAEASRLTEALRRYDDAYHHELAWWTAPS
MAV_2505|M.avium_104 DVGRDH-SVALDVLGPDARPGLVEAARLTEALRRYDDDYHHELDWWTSPS
MSMEG_3952|M.smegmatis_MC2_155 AAIDHE-AVRFVMLADDARGRLAEASQLTESLRRYDEFYHRELQWWTANV
TH_1331|M.thermoresistible__bu QNVFDD-TVMLTVLGDDARPRLAEASRLSESLRRYDASYHAELQWWTTPT
MUL_2417|M.ulcerans_Agy99 NTIDND-IAMLDVLTDEQRPQLVQASHLSAALRRDDLSYHAELDWWTSPF
. . : * *. *: *: * * *: ** **
Mb2058|M.bovis_AF2122/97 ETSEGIPHSSLVSAAESDRVTFGRDFPVVANTDRRPEFGHDRSKVLVLST
Rv2032|M.tuberculosis_H37Rv ETSEGIPHSSLVSAAESDRVTFGRDFPVVANTDRRPEFGHDRSKVLVLST
Mflv_3845|M.gilvum_PYR-GCK VATEGIPRSALVSAAEGDRVGIGRTFPVTTQPERRGELESDDAAIVVLST
Mvan_1383|M.vanbaalenii_PYR-1 AVAEGIPTGTLLSEAESDRVDIGRDFPAAGHRDRRGEIAQDHAKVVVLST
MAB_3903|M.abscessus_ATCC_1997 TLEDGVPRSALVSAADSDRVDIGRTFPVVAERASRPGLQEDQAKVLALST
MMAR_3487|M.marinum_M RESEGIPQSALASGTAAREVAVNRSFPVDWRSESSSAGRHDHATIVVLST
MAV_2505|M.avium_104 RKFDGIPESALVSEADDRRVDVNRRFPVDPLDERSSAGSYDDAKILVLST
MSMEG_3952|M.smegmatis_MC2_155 RHSDGVPAEALVSSPEAERVGVNRAFPSREHAERRPDVARDEARIAVLIT
TH_1331|M.thermoresistible__bu WVSQGLPPDALPSERERERVEIARAFPARGDSDRRPEVAEDRSTIVVLAT
MUL_2417|M.ulcerans_Agy99 VLTDGVPPLALASDTERQRVDLSRDFPVRSHQDRRPEITSDWSKILVLST
:*:* :* * .* . * ** * : : .* *
Mb2058|M.bovis_AF2122/97 YDNERASLLRCGEMLSAVLLDATMAGLATCTLTHITELHASRDLVAALIG
Rv2032|M.tuberculosis_H37Rv YDNERASLLRCGEMLSAVLLDATMAGLATCTLTHITELHASRDLVAALIG
Mflv_3845|M.gilvum_PYR-GCK EDDSRESVLRCGEALSAFLLDATMTGLATCTVTHITEVPAGRDVVASVLA
Mvan_1383|M.vanbaalenii_PYR-1 VDETRDGFVRCGEMLSSLLLDATMAGLATCVLSHLTEVPESRRIVESLTG
MAB_3903|M.abscessus_ATCC_1997 ARDTPKNLLRCGELLSEVLLEATLAGMATCTITHMLELTASREVVGAAIG
MMAR_3487|M.marinum_M PGDSRADALNCGAALSAVLLECTLAGLATCPVTHLTEMDAGRDTVRELTG
MAV_2505|M.avium_104 PEDTRADALRVGQVLSRILLECTAAGLATCPVTHVTELEAGRDLIRHLMA
MSMEG_3952|M.smegmatis_MC2_155 PEDTPRDAIACGEALSTILLECTMAGLATCPLTHITEMPESRRIILDLAG
TH_1331|M.thermoresistible__bu PEDSVRDALRCGEALSALLLECTMAGMATCPLTHMIEMQPSRDIVRELLD
MUL_2417|M.ulcerans_Agy99 PGDTRADVLRCGEVLSSVLLECTMAGMATCTLTHLIESNESRDIVADLIG
: . : * ** .**:.* :*:*** ::*: * .* :
Mb2058|M.bovis_AF2122/97 Q-PATPQALVRVGLAPEMEEPPPATPRRLIDEVFHVRAKDHR-
Rv2032|M.tuberculosis_H37Rv Q-PATPQALVRVGLAPEMEEPPPATPRRPIDEVFHVRAKDHR-
Mflv_3845|M.gilvum_PYR-GCK T-TSIPQVLVRVGAAPALDAIPAPTPRRPVHDVLEIRRGATC-
Mvan_1383|M.vanbaalenii_PYR-1 S-QAVPQVVVRVGSAPSGDAIGSPTPRRPVADVLEIRG-----
MAB_3903|M.abscessus_ATCC_1997 --RSHPQALIRVGLAPTFGRIPPPTPRRSLADVFEIRS-----
MMAR_3487|M.marinum_M G-QAEPQVLVRVGIAPTG-AAPELTPRRPLRDVLEIRR-----
MAV_2505|M.avium_104 DPAAVPQVLIRVGVEPEGESPPRPTPRRPLGDVLQFRPLSDR-
MSMEG_3952|M.smegmatis_MC2_155 T-TGIPQVLIRIGSAPPVDDVPPVTPRRSLAEVLEVDR-----
TH_1331|M.thermoresistible__bu R-PGLPQVLVRVGTTT-GGDAPPPTPRRPLAEVVEFRG-----
MUL_2417|M.ulcerans_Agy99 R-RGQPQVLIRAGITPLIEEIPAPTPRRGLDSIFEVRQKPEKG
. **.::* * . **** : .:...