For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GVCAAVVFGAPVWVAGVARAGDDVGFLVALQRIGITYPSPAQAIDAARAVCVCLDRGESGLAVVQEVTAR NPGFDMDAAAHFAVISATYYCPHHLAVT*
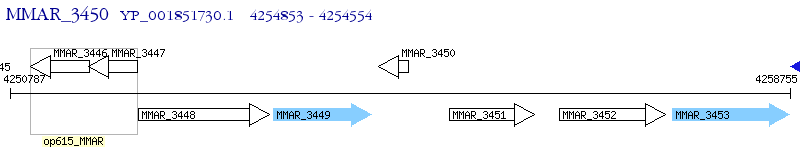
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3450 | - | - | 100% (99) | hypothetical protein MMAR_3450 |
| M. marinum M | MMAR_2686 | - | 4e-28 | 57.58% (99) | hypothetical protein MMAR_2686 |
| M. marinum M | MMAR_2049 | - | 1e-12 | 42.39% (92) | hypothetical protein MMAR_2049 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1833c | - | 8e-21 | 54.43% (79) | hypothetical protein Mb1833c |
| M. gilvum PYR-GCK | Mflv_5185 | - | 1e-10 | 42.22% (90) | hypothetical protein Mflv_5185 |
| M. tuberculosis H37Rv | Rv1804c | - | 8e-21 | 54.43% (79) | hypothetical protein Rv1804c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_2908 | - | 6e-23 | 48.00% (100) | hypothetical protein MAV_2908 |
| M. smegmatis MC2 155 | MSMEG_3681 | - | 6e-05 | 31.65% (79) | hypothetical protein MSMEG_3681 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2724 | - | 5e-53 | 97.98% (99) | hypothetical protein MUL_2724 |
| M. vanbaalenii PYR-1 | Mvan_1127 | - | 3e-09 | 34.07% (91) | hypothetical protein Mvan_1127 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3450|M.marinum_M -----------------------------------MGVCAAVVFGAPVWV
MUL_2724|M.ulcerans_Agy99 -----------------------------------MDVCAAVVFGAPVWV
Mb1833c|M.bovis_AF2122/97 -----------------------------------MRVVSTLLS-IPLMI
Rv1804c|M.tuberculosis_H37Rv -----------------------------------MRVVSTLLS-IPLMI
Mflv_5185|M.gilvum_PYR-GCK -----------------------------------MFARRAASAGIAAVA
Mvan_1127|M.vanbaalenii_PYR-1 MPDRHSVLRARTAVDVFAEVRFGLQRLDPPGIDMTTLTRLSAIAAMAAAA
MAV_2908|M.avium_104 -------------------------------MMKRLLAALGASAMIGATI
MSMEG_3681|M.smegmatis_MC2_155 ---------------------------MMAGLMGSMIGAAPALADPADPA
MMAR_3450|M.marinum_M -----AGVARA-------GDDVGFLVALQRIGITYPSPAQAIDAARAVCV
MUL_2724|M.ulcerans_Agy99 -----AGVARA-------GDDVGFLVALQRIGITYPSPAQAIDAARAVCV
Mb1833c|M.bovis_AF2122/97 GLAVPAHAGPS-------GDDAVFLASLERAGITYSHPDQAIASGKAVCA
Rv1804c|M.tuberculosis_H37Rv GLAVPAHAGPS-------GDDAVFLASLERAGITYSHPDQAIASGKAVCA
Mflv_5185|M.gilvum_PYR-GCK AVGLAAPASAS-------PEVAVFLDRLQKVGITSSNPYATIYDAYAVCR
Mvan_1127|M.vanbaalenii_PYR-1 AVALAASAAAA-------PQDAVFLDRLQKVGITSSNPYATVHGGYDVCR
MAV_2908|M.avium_104 GLAAPAHADPPPV---PDGDDGGFVAALQQAGFSFSSPGSAVAAGRAVCS
MSMEG_3681|M.smegmatis_MC2_155 VPGIPMDPAVPVPGAPGAPTDQAFVDALGQAGVNAVDPAQAAAMGEAVCP
. *: * : *.. * : . **
MMAR_3450|M.marinum_M CLDRGESGLAVVQEVTARNPGFDMDAAAHFAVISATYYCPHHLAVT----
MUL_2724|M.ulcerans_Agy99 CLDRGESGLAVVQEVTARNPGFDMDAAAHFAVISTTYYCPHHLAVT----
Mb1833c|M.bovis_AF2122/97 LVESGESGLQVVNELRTRNPGFSMDGCCKFAAISAHVYCPHQITKTSVSA
Rv1804c|M.tuberculosis_H37Rv LVESGESGLQVVNELRTRNPGFSMDGCCKFAAISAHVYCPHQITKTSVSA
Mflv_5185|M.gilvum_PYR-GCK ELDRGTSPTQVVGFVLGDNPDLDWEAAADYVVLANMTYCPPV--------
Mvan_1127|M.vanbaalenii_PYR-1 QLDLGTPHSQIVGFVLSDNPDLDWDSAADYVVLANMTYCPPV--------
MAV_2908|M.avium_104 CLNNGEPGLEVVHDVKTHNPGMDMEMASNFALLSAKYYCPHQLSKA----
MSMEG_3681|M.smegmatis_MC2_155 MLAEQGQSAADVASTVADTGGMPLGPATMFTGIAVSMFCPAMVSRLSQGV
: * . .: . :. :: :**
MMAR_3450|M.marinum_M ---------------
MUL_2724|M.ulcerans_Agy99 ---------------
Mb1833c|M.bovis_AF2122/97 K--------------
Rv1804c|M.tuberculosis_H37Rv K--------------
Mflv_5185|M.gilvum_PYR-GCK ---------------
Mvan_1127|M.vanbaalenii_PYR-1 ---------------
MAV_2908|M.avium_104 ---------------
MSMEG_3681|M.smegmatis_MC2_155 PPDLPMNLPWPMFGF