For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RVVSTLLSIPLMIGLAVPAHAGPSGDDAVFLASLERAGITYSHPDQAIASGKAVCALVESGESGLQVVNE LRTRNPGFSMDGCCKFAAISAHVYCPHQITKTSVSAK*
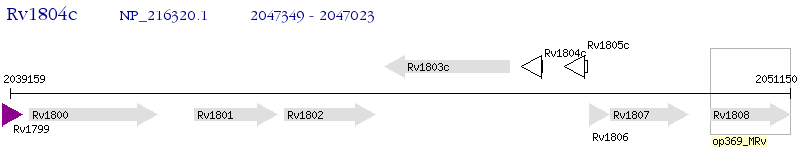
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1804c | - | - | 100% (108) | hypothetical protein Rv1804c |
| M. tuberculosis H37Rv | Rv1810 | - | 6e-26 | 53.06% (98) | hypothetical protein Rv1810 |
| M. tuberculosis H37Rv | Rv3067 | - | 4e-09 | 37.14% (70) | hypothetical protein Rv3067 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1833c | - | 2e-58 | 100.00% (108) | hypothetical protein Mb1833c |
| M. gilvum PYR-GCK | Mflv_5185 | - | 6e-10 | 34.38% (96) | hypothetical protein Mflv_5185 |
| M. leprae Br4923 | MLBr_00676 | - | 2e-05 | 30.21% (96) | hypothetical protein MLBr_00676 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2686 | - | 2e-27 | 51.92% (104) | hypothetical protein MMAR_2686 |
| M. avium 104 | MAV_2904 | - | 5e-26 | 56.32% (87) | hypothetical protein MAV_2904 |
| M. smegmatis MC2 155 | MSMEG_4689 | - | 8e-11 | 36.45% (107) | hypothetical protein MSMEG_4689 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2724 | - | 3e-20 | 53.16% (79) | hypothetical protein MUL_2724 |
| M. vanbaalenii PYR-1 | Mvan_1127 | - | 5e-09 | 28.72% (94) | hypothetical protein Mvan_1127 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5185|M.gilvum_PYR-GCK -------------------------------------MFARRAASAGIAA
Mvan_1127|M.vanbaalenii_PYR-1 --MPDRHSVLRARTAVDVFAEVRFGLQRLDPPGIDMTTLTRLSAIAAMAA
Rv1804c|M.tuberculosis_H37Rv ----------------------------------MRVVSTLLSI-PLMIG
Mb1833c|M.bovis_AF2122/97 ----------------------------------MRVVSTLLSI-PLMIG
MAV_2904|M.avium_104 -------------------------------------MLALLGIGAAALG
MMAR_2686|M.marinum_M ----------------------------------MKTLFALLGLSAMIW-
MUL_2724|M.ulcerans_Agy99 ----------------------------------MDVCAAVV-FGAPVW-
MSMEG_4689|M.smegmatis_MC2_155 ---------------------------MSARLPAVLVTAAALAAGVGATP
MLBr_00676|M.leprae_Br4923 MFLRLMLSALKALQRLGAVMNSLARIDHWIWLFRCQPLTIRLLVATAALF
Mflv_5185|M.gilvum_PYR-GCK VAAVGLAAPASASPEVAVFLDRLQKVGITSSNPYATIYDAYAVCRELDR-
Mvan_1127|M.vanbaalenii_PYR-1 AAAVALAASAAAAPQDAVFLDRLQKVGITSSNPYATVHGGYDVCRQLDL-
Rv1804c|M.tuberculosis_H37Rv LAVPAHAGPS---GDDAVFLASLERAGITYSHPDQAIASGKAVCALVES-
Mb1833c|M.bovis_AF2122/97 LAVPAHAGPS---GDDAVFLASLERAGITYSHPDQAIASGKAVCALVES-
MAV_2904|M.avium_104 YAAPARAEPQ---GDDAAFLASLDQSGITYSSSVQVIASAKAVCGLMGR-
MMAR_2686|M.marinum_M LAAPAHAHPDEGGGDDVGFLAALQNAGITYSNPDQAIGSAKAVCWCLDG-
MUL_2724|M.ulcerans_Agy99 VAGVARA------GDDVGFLVALQRIGITYPSPAQAIDAARAVCVCLDR-
MSMEG_4689|M.smegmatis_MC2_155 AHADPAAPGAPGNPVDRSFLTALNNAGVGYADPDAAVRLGQDVCPMLVEP
MLBr_00676|M.leprae_Br4923 TAATAFEVPAEADAIDDTFIKALNHAGVNFGEPRSAMTMGHYVCPILAKS
*: *:. *: . .: . ** :
Mflv_5185|M.gilvum_PYR-GCK GTSPTQVVGFVLGDNPDLDWEAAADYVVLANMTYCPPV------------
Mvan_1127|M.vanbaalenii_PYR-1 GTPHSQIVGFVLSDNPDLDWDSAADYVVLANMTYCPPV------------
Rv1804c|M.tuberculosis_H37Rv GESGLQVVNELRTRNPGFSMDGCCKFAAISAHVYCPHQITKTSVSAK---
Mb1833c|M.bovis_AF2122/97 GESGLQVVNELRTRNPGFSMDGCCKFAAISAHVYCPHQITKTSVSAK---
MAV_2904|M.avium_104 GETGLQVVTDIKDQNPGMTMDGAAQFAALASNAYCPHHLAPKGG------
MMAR_2686|M.marinum_M GESGLELVHDVKTHNPGFNMEAASQFAVLAATYYCPHHLSHA--------
MUL_2724|M.ulcerans_Agy99 GESGLAVVQEVTARNPGFDMDAAAHFAVISTTYYCPHHLAVT--------
MSMEG_4689|M.smegmatis_MC2_155 GKNFASVVTKLRGDRSVVSPDVAAFFAGIAISMYCPQMISGIGDGTLLRM
MLBr_00676|M.leprae_Br4923 GGNFAAAVQRIRG-NSDMSPQMAETFAKIAISIYCPTMMANVASGNLPSL
* * : .. . : . :. :: ***
Mflv_5185|M.gilvum_PYR-GCK -----------
Mvan_1127|M.vanbaalenii_PYR-1 -----------
Rv1804c|M.tuberculosis_H37Rv -----------
Mb1833c|M.bovis_AF2122/97 -----------
MAV_2904|M.avium_104 -----------
MMAR_2686|M.marinum_M -----------
MUL_2724|M.ulcerans_Agy99 -----------
MSMEG_4689|M.smegmatis_MC2_155 LAVPGLGALGR
MLBr_00676|M.leprae_Br4923 PPGPGIPGI--