For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAYEQPRHRARSPFWKSLAVFWLALSVAACSSSTTSAEGASTNANGTNNALEGVTVPDAFTPLTIAPISR PTFPFPGTDGKYHLAYDVQITNATGAPAALTSVDVVDAQDPSKVLTSFSGTQLVDPKCSYGNCNRLRQLT GRPSTDFTIPPQGSRALLLDFTFDKLAQFPKAVMLRLHGTGVTAPGSSAESGPLDTLGAPFDISAGTPRV ISPPLRGNNWVAFNGCCEPGWAHRDSILPLNMKLNNSQRFAIDWMRTNDQGNFFAGDKTNNDSYISYGSS VYAVADGTVTSTLDDMEANVPGILPASDPALAAKITVDNVDGNHIVLDIGGGAYAMYAHLIKGSLLVKPG DKVKKGQEIAKLGNTGNSNAPHLHFQLMDGPSLLEADALPYVLDTFSYQGQVSPATVWDADNYLSGSFFG PDRLAAPEPRTKQLPVLLAIVNFG
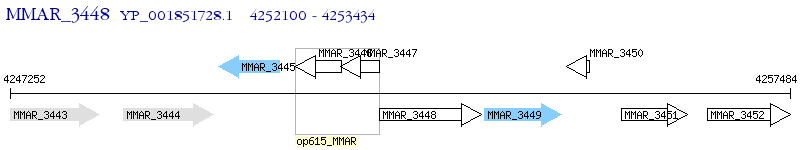
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3448 | - | - | 100% (444) | hypothetical protein MMAR_3448 |
| M. marinum M | MMAR_4551 | - | 9e-06 | 31.43% (105) | hypothetical protein MMAR_4551 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0975c | - | 6e-05 | 38.78% (49) | hypothetical protein Mb0975c |
| M. gilvum PYR-GCK | Mflv_1864 | - | 1e-09 | 37.36% (91) | peptidase M23B |
| M. tuberculosis H37Rv | Rv0950c | - | 6e-05 | 38.78% (49) | hypothetical protein Rv0950c |
| M. leprae Br4923 | MLBr_00154 | - | 2e-05 | 33.75% (80) | hypothetical protein MLBr_00154 |
| M. abscessus ATCC 19977 | MAB_1055c | - | 2e-05 | 41.18% (51) | hypothetical protein MAB_1055c |
| M. avium 104 | MAV_1073 | - | 4e-08 | 33.33% (105) | M23 peptidase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_0247 | - | 2e-61 | 35.73% (389) | secreted peptidase |
| M. thermoresistible (build 8) | TH_1774 | - | 7e-09 | 48.28% (58) | peptidoglycan-binding LysM |
| M. ulcerans Agy99 | MUL_2722 | - | 0.0 | 98.92% (369) | hypothetical protein MUL_2722 |
| M. vanbaalenii PYR-1 | Mvan_4871 | - | 2e-08 | 36.26% (91) | peptidase M23B |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0975c|M.bovis_AF2122/97 ----MAAIRTPR----DRWPHHHRN--------EVTEIIPLDGF------
Rv0950c|M.tuberculosis_H37Rv ----MAAIRTPR----DRWPHHHRN--------EVTEIIPLDGF------
MAV_1073|M.avium_104 ----MRVGRAPG----ARSAEPHRT--------EVTEILPLDGFD-----
MLBr_00154|M.leprae_Br4923 --MAAEAVRSFR----NRWLNHQRN--------DVTEIIPVDGF------
Mflv_1864|M.gilvum_PYR-GCK ----MPQHRLPR----SRAALTTPG------LSEVTDIIPFDEFGA----
Mvan_4871|M.vanbaalenii_PYR-1 ----MPQHRLPR----SIEVLAAPG------PTEVTDIIPFNEFG-----
TH_1774|M.thermoresistible__bu ----LAKHRAVSPAGDSTDAHRHPRQGFDPHPAEITEIIPLSGFGR----
MAB_1055c|M.abscessus_ATCC_199 ----MPIDEFGL-----SDALERPE----LEDYDVYEDAPEEGA------
MMAR_3448|M.marinum_M MAYEQPRHRARSPFWKSLAVFWLALSVAACSSSTTSAEGASTNANGTNNA
MUL_2722|M.ulcerans_Agy99 MLIEYHWRRRRFDQRQ------RHRQRARGSNGSRRVHTADHRAN-----
MSMEG_0247|M.smegmatis_MC2_155 --MRYLTLAVSLGTATALAVLTACSSAETTESESTTTTTPKVQPT-----
.
Mb0975c|M.bovis_AF2122/97 -------------LDGLALYDELDFAELDDLDLG-DDCVFDYEAQLLAAP
Rv0950c|M.tuberculosis_H37Rv -------------LDGLALYDELDFAELDDLDLG-DDCVFDYEAQLLAAP
MAV_1073|M.avium_104 -------------FDDLDFCDDAG--DHGDLDFS-TDSPFDNEAQVLLAP
MLBr_00154|M.leprae_Br4923 --------------DDFDDLYDLDFAEIDELQFSDDDYNFCNETQVLLAS
Mflv_1864|M.gilvum_PYR-GCK -------------LGDIDDLEFLQ------------DAAFGRDAEVIDAP
Mvan_4871|M.vanbaalenii_PYR-1 ---------------DLSDLEFLP------------HAAFDRDTQVIHAP
TH_1774|M.thermoresistible__bu -------------LSDLGADTDFYGDAWDD----EWDVAFDRESEVVSAP
MAB_1055c|M.abscessus_ATCC_199 ---------------EIEDQTALD------------QLEQDWEED---TD
MMAR_3448|M.marinum_M LEGVTVPDAFTPLTIAPISRPTFPFPGTDGKYHLAYDVQITNATGAPAAL
MUL_2722|M.ulcerans_Agy99 ----------QPADVS--------VPGTDDKYHLAYDVQITNATGAPAAL
MSMEG_0247|M.smegmatis_MC2_155 ------------PVIGSVLAEPIPVPTTDGRTHLAYELMLTNPMSGNVTL
. :
Mb0975c|M.bovis_AF2122/97 ELDDLDDADDLAPEWLVAPTVVLTP-------------------------
Rv0950c|M.tuberculosis_H37Rv ELDDLDDADDLAPEWLVAPTVVLTP-------------------------
MAV_1073|M.avium_104 ELDDLNETDDLRPLWLAAPVTEVLPR--------TTVAARTERRPERPRG
MLBr_00154|M.leprae_Br4923 EIDDLQDIDSLASEPLAASANVAPN-------------------------
Mflv_1864|M.gilvum_PYR-GCK ELDDCHDTDDLTPLRLAVPAEFRAEPAAARRASTRAYRDSHTDVSDGSAK
Mvan_4871|M.vanbaalenii_PYR-1 ELDDLHDTDDLTPLQLAVPSEFRAAPRADRRPSGRAYRDSHTDVSDGTAK
TH_1774|M.thermoresistible__bu ELDDL-DPDAEAPLRLVVPSEFRPDPRDER-PSGHAYRDSHTDVSDGTAA
MAB_1055c|M.abscessus_ATCC_199 EADRIRASIEATADWFAASAARTRDP--------------------RDTP
MMAR_3448|M.marinum_M TSVDVVDAQDPSKVLTSFSGTQLVDP--------------------KCSY
MUL_2722|M.ulcerans_Agy99 TSVDVVDSQDPSKVLTSFSGTQLVDP--------------------KCSY
MSMEG_0247|M.smegmatis_MC2_155 NAVSVMDGD---RKLLTLSGDSLKYW--------------------TRAL
.
Mb0975c|M.bovis_AF2122/97 -EVTPVSRRVGQHRK-QPIGAARGRLLISAMAAGAAAAAAHTAIQQ--SE
Rv0950c|M.tuberculosis_H37Rv -EVTPVSRRVGQHRK-QPIGAARGRLLISAMAAGAAAAAAHTAIQQ--SE
MAV_1073|M.avium_104 ADVVGVARRGGQHRK-QPTSAARGRLLISAMAAGAAAAAAHTATSH--AD
MLBr_00154|M.leprae_Br4923 --FTSAPRRPCQHRK-QPTSATRGRLLISAMAAGAMATAAHTVISH--DD
Mflv_1864|M.gilvum_PYR-GCK TDVIDMRAHKGQHRKTEVP--VKGRLVVAAMAVGATAAGAYTMTKN--VP
Mvan_4871|M.vanbaalenii_PYR-1 TDIIDMRGRTGLHRKAEVP--VKGRLVVAAMAVGATAAGAYTMTNG--TA
TH_1774|M.thermoresistible__bu TDVIAI-GRRGQHRKPEPTSAGKGRLMIAAMAAGATAAGAYSVVNGDDEK
MAB_1055c|M.abscessus_ATCC_199 TDKLPRISSGGVHRRREIGQVGKTRLALAAMAAGAAAAAGYNALSE----
MMAR_3448|M.marinum_M GNCNRLRQLTGRPSTDFTIPPQGSRALLLDFTFDKLAQFPKAVMLRLHGT
MUL_2722|M.ulcerans_Agy99 GNCNRLRQLTGRPSTDFTIPPQGSRALLLDFTFDKLAQFPKAVMLRLHGT
MSMEG_0247|M.smegmatis_MC2_155 GNPAAPANVLG---------PGQSATVWLDVVADDPAQVPTELTHAIDLT
.. . *
Mb0975c|M.bovis_AF2122/97 TPRTETVLTAHASALNEGSG-SNPPRGVQVIAAQPAASAAVHNAEFARGV
Rv0950c|M.tuberculosis_H37Rv TPRTETVLTAHASALNEGSG-SNPPRGVQVIAAQPAASAAVHNAEFARGV
MAV_1073|M.avium_104 TPKTDAVLTANASSLTGGAGGSNTVRGPQVIAAEPAATAALHNQELAKGV
MLBr_00154|M.leprae_Br4923 IPKTETVLTANASALS-GATVSDATQGVQVITVHPAASTAVHNEEHAKGT
Mflv_1864|M.gilvum_PYR-GCK AAPAETVLASDSTTLGDGTVITGSVDGMQIVSVAPASNSTVHAEEITKAA
Mvan_4871|M.vanbaalenii_PYR-1 KAPAETVLASDHTTLGDSTVIAGSTDGMQIVTVAPAANSTVHAEEITKAA
TH_1774|M.thermoresistible__bu SSTDATVLAADQAAMPD--AIDGATGGMQIVTVAQPAKAIVHAEEITKAA
MAB_1055c|M.abscessus_ATCC_199 -HDAATAADHHGLALGN-SAAVMATHGPQLVSAPLATDASVTDEQLAKAT
MMAR_3448|M.marinum_M GVTAPGSSAESGPLDTLGAPFDISAGTPRVISPPLRGNNWVAFNGCCEPG
MUL_2722|M.ulcerans_Agy99 GVTAPGSSAESGPLDTLGAPFDISAGTPRVIGPPLRGNNWVAFNGCCEPG
MSMEG_0247|M.smegmatis_MC2_155 -VAKPMPPLIPANVTETIAPVSVQTRKPPSISPPLTGDRWLDINSCCD-M
: :
Mb0975c|M.bovis_AF2122/97 AFAEERAEREARLQRPLYVMPTKGIFTSSFGYRWGVLHAGIDLANAIGTP
Rv0950c|M.tuberculosis_H37Rv AFAEERAEREARLQRPLYVMPTKGIFTSSFGYRWGVLHAGIDLANAIGTP
MAV_1073|M.avium_104 AFANDRAQREARLQQPLYVMPTKGIFTSNFGYRWGVLHAGIDLANSIGTP
MLBr_00154|M.leprae_Br4923 AFAHERAQREARLSQPLYVMPTKGIFTSNFGYRWGVLHAGIDLANAIGTP
Mflv_1864|M.gilvum_PYR-GCK AFAQERAEREARLTKPQFVMPTTGVWTSGFGYRWGVLHAGIDIANAIGTP
Mvan_4871|M.vanbaalenii_PYR-1 AFAQERAEREARLTKPQFAMPTTGVWTSGFGYRWGVLHAGIDIANAIGTP
TH_1774|M.thermoresistible__bu AFAQERAEREARLARPQFVMPTTGVWTSGFGYRWGVLHGGIDIANSVGTP
MAB_1055c|M.abscessus_ATCC_199 AFATERADREKRLLAPRFVMPTNGTFTSGFGYRWGALHGGIDIANSIGTP
MMAR_3448|M.marinum_M WAHRDSILPLNMKLNNSQRFAIDWMRTNDQGNFFAGDKTNNDSYISYGSS
MUL_2722|M.ulcerans_Agy99 WAHRDSILPLNMKLNNSQRFAIDWMRTNDQGNFFAGDKTNNDSYISYGSS
MSMEG_0247|M.smegmatis_MC2_155 TAHRMALNPINGKIWAAERFAIDYVQLGTDGRIYTGNKAAVESYPYFGAE
:. . * : : : *:
Mb0975c|M.bovis_AF2122/97 IYAVSDGVVIDAGPTAG-----------------------YGMWVKLLHA
Rv0950c|M.tuberculosis_H37Rv IYAVSDGVVIDAGPTAG-----------------------YGMWVKLLHA
MAV_1073|M.avium_104 IYAVSDGVVIEAGPAGG-----------------------YGMLVKLRHA
MLBr_00154|M.leprae_Br4923 ILAVSDGVVIDVGPTAG-----------------------YGMWVKLRHA
Mflv_1864|M.gilvum_PYR-GCK VLAAADGIVIASGAEGG-----------------------YGNAVKLRHA
Mvan_4871|M.vanbaalenii_PYR-1 ILAAADGVVIAAGPEGG-----------------------YGNLVKLRHA
TH_1774|M.thermoresistible__bu ILAAADGVVIETGPQGG-----------------------YGIWVKLRHY
MAB_1055c|M.abscessus_ATCC_199 IVAAADGVVIATGPTAG-----------------------YGAWVKIRHS
MMAR_3448|M.marinum_M VYAVADGTVTSTLDDMEANVPGILPASDPALAAKITVDNVDGNHIVLDIG
MUL_2722|M.ulcerans_Agy99 VYAVADGTVTSTLDDMEANVPGILPASDPALAAKITVDNVDGNHIVLDIG
MSMEG_0247|M.smegmatis_MC2_155 IRAVADGPVVSVLDGQPEQTPGVTPSG-------LTLEQYGGNHIVQDIG
: *.:** * * :
Mb0975c|M.bovis_AF2122/97 DGTVTLYGHVN--TTLVSVGERVMAGDQIATMGSRGFSTGPHLHFEVLLG
Rv0950c|M.tuberculosis_H37Rv DGTVTLYGHVN--TTLVSVGERVMAGDQIATMGSRGFSTGPHLHFEVLLG
MAV_1073|M.avium_104 DGTVTLYGHIN--TALVSVGERVMAGDQIATMGNRGNSTGPHLHFEVLQG
MLBr_00154|M.leprae_Br4923 DGTVTLYGHVN--TTLVNVGQYVVAGDQIATMGTRGNSTGPHLHFEVLLS
Mflv_1864|M.gilvum_PYR-GCK DGTVTLYGHNS--SLLVSVGERVMAGDQIAKMGNTGNSTGPHLHFEVHLN
Mvan_4871|M.vanbaalenii_PYR-1 DGTVTLYGHNS--SVLVNVGERVMAGDQIAKMGNTGNSTGPHCHFEVHLN
TH_1774|M.thermoresistible__bu DGTVTVYAHLS--SVLVNVGQRVMAGDQIARMGNTGNSTGPHLHFEVLLD
MAB_1055c|M.abscessus_ATCC_199 DGTVTLYGHIN--TWEVSVGQRVMAGDRIATIGNRGNSTGPHLHFEVLLG
MMAR_3448|M.marinum_M GGAYAMYAHLIKGSLLVKPGDKVKKGQEIAKLGNTGNSNAPHLHFQLMDG
MUL_2722|M.ulcerans_Agy99 GGAYAMYAHLIKGSLLVKPGDKVKKGQEIAKLGNTGNSNAPHLHFQLMDG
MSMEG_0247|M.smegmatis_MC2_155 DGVYAFYAHLQPGSLKVKPGDQLSTGQAIASLGNSGNSDAPHLHFHVMDG
.*. :.*.* : *. *: : *: ** :*. * * .** **.: .
Mb0975c|M.bovis_AF2122/97 GTERVDPVPWLAKRGLSV-GNYTG--------------------------
Rv0950c|M.tuberculosis_H37Rv GTERVDPVPWLAKRGLSV-GNYTG--------------------------
MAV_1073|M.avium_104 GTERIDPVPWLAKRGLMV-GNYAG--------------------------
MLBr_00154|M.leprae_Br4923 GSERIDPVPWLAKRGIYV-GNYTG--------------------------
Mflv_1864|M.gilvum_PYR-GCK GTDRVDPTGWLAKRGLSP-GGYVG--------------------------
Mvan_4871|M.vanbaalenii_PYR-1 GTDRVDPVGWLAKRGLTP-GGYVG--------------------------
TH_1774|M.thermoresistible__bu GVNRVDPAAWLAKRGVGPNSGYMR--------------------------
MAB_1055c|M.abscessus_ATCC_199 GSQRIDPQGWLANKGLTF-TRFGD--------------------------
MMAR_3448|M.marinum_M PSLLEADALPYVLDTFSYQGQVSPATVWDADNYLSGSFFGPDRLAAPEPR
MUL_2722|M.ulcerans_Agy99 PSLLEADALPYVLDTFSYQGQISPATVWDADNYLSGSFFGPDRLAAPEPR
MSMEG_0247|M.smegmatis_MC2_155 PDPLAADGLPFVFDSFRLDSRLTSATAGDALLDGKPAALQPG--FATREV
. .
Mb0975c|M.bovis_AF2122/97 ----------------
Rv0950c|M.tuberculosis_H37Rv ----------------
MAV_1073|M.avium_104 ----------------
MLBr_00154|M.leprae_Br4923 ----------------
Mflv_1864|M.gilvum_PYR-GCK ----------------
Mvan_4871|M.vanbaalenii_PYR-1 ----------------
TH_1774|M.thermoresistible__bu ----------------
MAB_1055c|M.abscessus_ATCC_199 ----------------
MMAR_3448|M.marinum_M TKQLPVLLAIVNFG--
MUL_2722|M.ulcerans_Agy99 TKQLPVLLAIVNFG--
MSMEG_0247|M.smegmatis_MC2_155 SEVSPLVLDVMTYASD