For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PIDEFGLSDALERPELEDYDVYEDAPEEGAEIEDQTALDQLEQDWEEDTDEADRIRASIEATADWFAASA ARTRDPRDTPTDKLPRISSGGVHRRREIGQVGKTRLALAAMAAGAAAAAGYNALSEHDAATAADHHGLAL GNSAAVMATHGPQLVSAPLATDASVTDEQLAKATAFATERADREKRLLAPRFVMPTNGTFTSGFGYRWGA LHGGIDIANSIGTPIVAAADGVVIATGPTAGYGAWVKIRHSDGTVTLYGHINTWEVSVGQRVMAGDRIAT IGNRGNSTGPHLHFEVLLGGSQRIDPQGWLANKGLTFTRFGD*
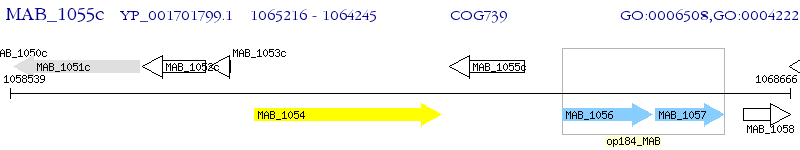
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1055c | - | - | 100% (323) | hypothetical protein MAB_1055c |
| M. abscessus ATCC 19977 | MAB_3197 | - | 6e-06 | 35.45% (110) | hypothetical protein MAB_3197 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0975c | - | 6e-75 | 48.00% (325) | hypothetical protein Mb0975c |
| M. gilvum PYR-GCK | Mflv_1864 | - | 2e-73 | 47.62% (336) | peptidase M23B |
| M. tuberculosis H37Rv | Rv0950c | - | 7e-75 | 48.00% (325) | hypothetical protein Rv0950c |
| M. leprae Br4923 | MLBr_00154 | - | 2e-70 | 46.91% (307) | hypothetical protein MLBr_00154 |
| M. marinum M | MMAR_4551 | - | 5e-74 | 47.53% (324) | hypothetical protein MMAR_4551 |
| M. avium 104 | MAV_1073 | - | 2e-72 | 48.61% (323) | M23 peptidase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_5526 | - | 4e-77 | 50.00% (344) | peptidoglycan-binding LysM |
| M. thermoresistible (build 8) | TH_1774 | - | 5e-73 | 48.72% (312) | peptidoglycan-binding LysM |
| M. ulcerans Agy99 | MUL_4721 | - | 4e-73 | 47.53% (324) | hypothetical protein MUL_4721 |
| M. vanbaalenii PYR-1 | Mvan_4871 | - | 5e-70 | 45.75% (341) | peptidase M23B |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0975c|M.bovis_AF2122/97 ------------MAAIRTPRDRWPHHHRNEVTEIIPLDGFLDGLALYDEL
Rv0950c|M.tuberculosis_H37Rv ------------MAAIRTPRDRWPHHHRNEVTEIIPLDGFLDGLALYDEL
MMAR_4551|M.marinum_M MSQHRYAR-TAVVGDPRTSRDRWLQHHRNEVTEIIPLDGFDE----LDEL
MUL_4721|M.ulcerans_Agy99 MSQHRYAR-TAVVGNPRTSRDRWLQHHRNEVTEIIPLDGF-------DEL
MAV_1073|M.avium_104 ------------MRVGRAPGARSAEPHRTEVTEILPLDGFD-----FDDL
MLBr_00154|M.leprae_Br4923 ----------MAAEAVRSFRNRWLNHQRNDVTEIIPVDGFDD-FDDLYDL
Mflv_1864|M.gilvum_PYR-GCK MPQHRLPR----SRAALTTPG------LSEVTDIIPFDEFGA------LG
Mvan_4871|M.vanbaalenii_PYR-1 MPQHRLPR----SIEVLAAPG------PTEVTDIIPFNEFG---------
TH_1774|M.thermoresistible__bu LAKHRAVSPAGDSTDAHRHPRQGFDPHPAEITEIIPLSGFGR------LS
MSMEG_5526|M.smegmatis_MC2_155 ------------------------------MTDIIPFSEFSK------AF
MAB_1055c|M.abscessus_ATCC_199 ----------------------------------MPIDEFGLS----DAL
:*.. *
Mb0975c|M.bovis_AF2122/97 DFAELD----DLDLGDDC-VFDYEAQLLAAPELDDLDDADDLAPEWLVAP
Rv0950c|M.tuberculosis_H37Rv DFAELD----DLDLGDDC-VFDYEAQLLAAPELDDLDDADDLAPEWLVAP
MMAR_4551|M.marinum_M ELADLD----DLDFGDP--EFDTGRQLLQAPELDDLDEIDNLIPAWLATP
MUL_4721|M.ulcerans_Agy99 ELADLD----DLDFGDP--EFDTGRQLLQAPELDDLDEIDNLIPAWLATP
MAV_1073|M.avium_104 DFCDDAGDHGDLDFSTDS-PFDNEAQVLLAPELDDLNETDDLRPLWLAAP
MLBr_00154|M.leprae_Br4923 DFAEID----ELQFSDDDYNFCNETQVLLASEIDDLQDIDSLASEPLAAS
Mflv_1864|M.gilvum_PYR-GCK DIDDLEFLQ--------DAAFGRDAEVIDAPELDDCHDTDDLTPLRLAVP
Mvan_4871|M.vanbaalenii_PYR-1 DLSDLEFLP--------HAAFDRDTQVIHAPELDDLHDTDDLTPLQLAVP
TH_1774|M.thermoresistible__bu DLGADTDFYGDAWDDEWDVAFDRESEVVSAPELDDL-DPDAEAPLRLVVP
MSMEG_5526|M.smegmatis_MC2_155 DQFAALDTQISSAYDN-LSAFEREARVIEAPELDDLNDTDDLEPVRLDIP
MAB_1055c|M.abscessus_ATCC_199 ERPELEDYDVYEDAPEEGAEIEDQT-ALDQLEQDWEEDTDEADRIRASIE
: : : * * : *
Mb0975c|M.bovis_AF2122/97 -TVVLT----------------------------------PEVTPVSRRV
Rv0950c|M.tuberculosis_H37Rv -TVVLT----------------------------------PEVTPVSRRV
MMAR_4551|M.marinum_M -SPVLVGAT-----------------SEALPEPSAAVPPAANVAPAPRRG
MUL_4721|M.ulcerans_Agy99 -SPVLVGAT-----------------SEALPEPSAAVPPAANVAPAPRRG
MAV_1073|M.avium_104 VTEVLPRTT-----------------VAARTERRPERPRGADVVGVARRG
MLBr_00154|M.leprae_Br4923 -ANVAP-----------------------------------NFTSAPRRP
Mflv_1864|M.gilvum_PYR-GCK -AEFRAEPA--------AARRASTRAYRDSHTDVSDGSAKTDVIDMRAHK
Mvan_4871|M.vanbaalenii_PYR-1 -SEFRAAPR--------ADRRPSGRAYRDSHTDVSDGTAKTDIIDMRGRT
TH_1774|M.thermoresistible__bu -SEFRPDPR--------DER-PSGHAYRDSHTDVSDGTAATDVIAI-GRR
MSMEG_5526|M.smegmatis_MC2_155 -SEFLADARDAGERSASRRKRNATHAYRDSHTDVSDGTAATDVIDMTGRR
MAB_1055c|M.abscessus_ATCC_199 ATADWFAAS--------------------AARTRDPRDTPTDKLPRISSG
: :
Mb0975c|M.bovis_AF2122/97 GQHRK-QPIGAARGRLLISAMAAGAAAAAAHTAIQQ--SETPRTETVLTA
Rv0950c|M.tuberculosis_H37Rv GQHRK-QPIGAARGRLLISAMAAGAAAAAAHTAIQQ--SETPRTETVLTA
MMAR_4551|M.marinum_M GQHRK-QSTSAAKGRLLIGSMAAGAAAAAAHTVTHQ--SDVTSMDTVLTA
MUL_4721|M.ulcerans_Agy99 GQHRK-QSTSAVKGRLLIGSMAAGAAAAAAHTVTHQ--SDVTSMDTVLTA
MAV_1073|M.avium_104 GQHRK-QPTSAARGRLLISAMAAGAAAAAAHTATSH--ADTPKTDAVLTA
MLBr_00154|M.leprae_Br4923 CQHRK-QPTSATRGRLLISAMAAGAMATAAHTVISH--DDIPKTETVLTA
Mflv_1864|M.gilvum_PYR-GCK GQHRKTEVP--VKGRLVVAAMAVGATAAGAYTMTKN--VPAAPAETVLAS
Mvan_4871|M.vanbaalenii_PYR-1 GLHRKAEVP--VKGRLVVAAMAVGATAAGAYTMTNG--TAKAPAETVLAS
TH_1774|M.thermoresistible__bu GQHRKPEPTSAGKGRLMIAAMAAGATAAGAYSVVNGDDEKSSTDATVLAA
MSMEG_5526|M.smegmatis_MC2_155 GAHRK-ETTGALKSRLMIAAVAAGATAAGAYSFGNG-TEAQAADDTVLAT
MAB_1055c|M.abscessus_ATCC_199 GVHRRREIGQVGKTRLALAAMAAGAAAAAGYNALSEHDAATAADHHGLAL
**: : : ** :.::*.** *:..:. . *:
Mb0975c|M.bovis_AF2122/97 HASALNEGSG-SNPPRGVQVIAAQPAASAAVHNAEFARGVAFAEERAERE
Rv0950c|M.tuberculosis_H37Rv HASALNEGSG-SNPPRGVQVIAAQPAASAAVHNAEFARGVAFAEERAERE
MMAR_4551|M.marinum_M NASALIGGSD-SNEARGVQVIAAQPATNVEAHKAEFARGVAFAQERAQRE
MUL_4721|M.ulcerans_Agy99 NASALIGGSD-SNEARGVQVIAAQPATNVEAHKAEFARGVAFAQERAQRE
MAV_1073|M.avium_104 NASSLTGGAGGSNTVRGPQVIAAEPAATAALHNQELAKGVAFANDRAQRE
MLBr_00154|M.leprae_Br4923 NASALS-GATVSDATQGVQVITVHPAASTAVHNEEHAKGTAFAHERAQRE
Mflv_1864|M.gilvum_PYR-GCK DSTTLGDGTVITGSVDGMQIVSVAPASNSTVHAEEITKAAAFAQERAERE
Mvan_4871|M.vanbaalenii_PYR-1 DHTTLGDSTVIAGSTDGMQIVTVAPAANSTVHAEEITKAAAFAQERAERE
TH_1774|M.thermoresistible__bu DQAAMPD--AIDGATGGMQIVTVAQPAKAIVHAEEITKAAAFAQERAERE
MSMEG_5526|M.smegmatis_MC2_155 GQTTVDG-ASVSGSNDGMQIVSVASNISSAVHAEEITKAAAFAQERAERE
MAB_1055c|M.abscessus_ATCC_199 GN------SAAVMATHGPQLVSAPLATDASVTDEQLAKATAFATERADRE
* *:::. : ::..*** :**:**
Mb0975c|M.bovis_AF2122/97 ARLQRPLYVMPTKGIFTSSFGYRWGVLHAGIDLANAIGTPIYAVSDGVVI
Rv0950c|M.tuberculosis_H37Rv ARLQRPLYVMPTKGIFTSSFGYRWGVLHAGIDLANAIGTPIYAVSDGVVI
MMAR_4551|M.marinum_M ARLQQPLYVMPTKGIFTSNFGYRWGVLHAGIDLANSIGTPIYAVSDGLVI
MUL_4721|M.ulcerans_Agy99 ARLQQPLYVMPTKGIFTSNFGYRWGVLHAGIDLANSIGTPIYAVSDGLVI
MAV_1073|M.avium_104 ARLQQPLYVMPTKGIFTSNFGYRWGVLHAGIDLANSIGTPIYAVSDGVVI
MLBr_00154|M.leprae_Br4923 ARLSQPLYVMPTKGIFTSNFGYRWGVLHAGIDLANAIGTPILAVSDGVVI
Mflv_1864|M.gilvum_PYR-GCK ARLTKPQFVMPTTGVWTSGFGYRWGVLHAGIDIANAIGTPVLAAADGIVI
Mvan_4871|M.vanbaalenii_PYR-1 ARLTKPQFAMPTTGVWTSGFGYRWGVLHAGIDIANAIGTPILAAADGVVI
TH_1774|M.thermoresistible__bu ARLARPQFVMPTTGVWTSGFGYRWGVLHGGIDIANSVGTPILAAADGVVI
MSMEG_5526|M.smegmatis_MC2_155 ARLSRPLFVMPTKGVWTSGFGYRWGVLHGGIDIANSIGTPIYAASDGVVT
MAB_1055c|M.abscessus_ATCC_199 KRLLAPRFVMPTNGTFTSGFGYRWGALHGGIDIANSIGTPIVAAADGVVI
** * :.***.* :**.******.**.***:**::***: *.:**:*
Mb0975c|M.bovis_AF2122/97 DAGPTAGYGMWVKLLHADGTVTLYGHVNTTLVSVGERVMAGDQIATMGSR
Rv0950c|M.tuberculosis_H37Rv DAGPTAGYGMWVKLLHADGTVTLYGHVNTTLVSVGERVMAGDQIATMGSR
MMAR_4551|M.marinum_M DAGPTAGYGMWVKIRHADGTVTLYGHVNTTLVSVGQRVMAGDQIATMGNR
MUL_4721|M.ulcerans_Agy99 DAGPTAGYGMWVKIRHADGTVTLYGHVNTTLVNVGQRVMAGDHIATMGNR
MAV_1073|M.avium_104 EAGPAGGYGMLVKLRHADGTVTLYGHINTALVSVGERVMAGDQIATMGNR
MLBr_00154|M.leprae_Br4923 DVGPTAGYGMWVKLRHADGTVTLYGHVNTTLVNVGQYVVAGDQIATMGTR
Mflv_1864|M.gilvum_PYR-GCK ASGAEGGYGNAVKLRHADGTVTLYGHNSSLLVSVGERVMAGDQIAKMGNT
Mvan_4871|M.vanbaalenii_PYR-1 AAGPEGGYGNLVKLRHADGTVTLYGHNSSVLVNVGERVMAGDQIAKMGNT
TH_1774|M.thermoresistible__bu ETGPQGGYGIWVKLRHYDGTVTVYAHLSSVLVNVGQRVMAGDQIARMGNT
MSMEG_5526|M.smegmatis_MC2_155 DVGPTAGYGAWVKIRHSDGTVTLYGHINTWLVSVGERVMAGDQIATMGNR
MAB_1055c|M.abscessus_ATCC_199 ATGPTAGYGAWVKIRHSDGTVTLYGHINTWEVSVGQRVMAGDRIATIGNR
*. .*** **: * *****:*.* .: *.**: *:***:** :*.
Mb0975c|M.bovis_AF2122/97 GFSTGPHLHFEVLLGGTERVDPVPWLAKRGLSV-GNYTG
Rv0950c|M.tuberculosis_H37Rv GFSTGPHLHFEVLLGGTERVDPVPWLAKRGLSV-GNYTG
MMAR_4551|M.marinum_M GFSTGPHLHFEVLQGGTERIDPVPWLAKRGLNV-GNYAG
MUL_4721|M.ulcerans_Agy99 GFSTGPHLHFEVLQGGTERIDPVPWLAKRGLNV-GNYAG
MAV_1073|M.avium_104 GNSTGPHLHFEVLQGGTERIDPVPWLAKRGLMV-GNYAG
MLBr_00154|M.leprae_Br4923 GNSTGPHLHFEVLLSGSERIDPVPWLAKRGIYV-GNYTG
Mflv_1864|M.gilvum_PYR-GCK GNSTGPHLHFEVHLNGTDRVDPTGWLAKRGLSP-GGYVG
Mvan_4871|M.vanbaalenii_PYR-1 GNSTGPHCHFEVHLNGTDRVDPVGWLAKRGLTP-GGYVG
TH_1774|M.thermoresistible__bu GNSTGPHLHFEVLLDGVNRVDPAAWLAKRGVGPNSGYMR
MSMEG_5526|M.smegmatis_MC2_155 GYSTGPHLHFEVLTNGTNRIDPVGWLAKRGLSP-GSYVG
MAB_1055c|M.abscessus_ATCC_199 GNSTGPHLHFEVLLGGSQRIDPQGWLANKGLTF-TRFGD
* ***** **** .* :*:** ***::*: :