For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVPEQSPGDVPEVAAPAPEPAPEDVTPLPDLLHGRHRAGPDRERRPVYVDLLPPCNAGCPAGENIQAWLA HARVGENERAWRQLTADNPFAAIHGRVCYHPCESVCNRADLDASVSIHAVERFLGDTAREKQWRFQTAPQ PTGKRVLVVGAGPSGLSAAYHLARLGHHVEVRDAGAEPGGMMRYGIPSYRLPRDVLDAEIERIATLGVQL TTGHRVEDLAEERETGNFDAVFVAVGAHLAKRVNIPSRDARSLIDAVSFLHQVAAGEKPELGSHVAVYGG GNTAMDAARVARRLGAQDTVIIYRRTEAQMPAHEEERQDAQREGVQINWLRTITAFDDHEIRVEQMELDD TGRPRPTGRFETLAADTVIMALGQETESAFMRTLPGVEFDRDGSVRVSPTLMTGCPGVFAGGDMVPCERT VTVGVGHGKKAAHYIDAWLRGDPTQRPPKHPVADFDRLHLWYFGDARRRQQPELSPEDRISGFDEVMGGL SAEAATFEAGRCLSCGNCFECDGCLGACPEDAVIKLGRGHRYRFDYDRCTGCATCYDQCPVHAIEMIAEP R
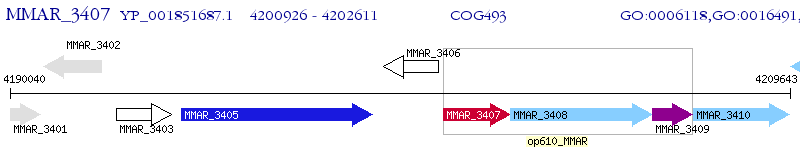
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3407 | gltD_1 | - | 100% (561) | NADH-dependent glutamate synthase (small subunit) GltD |
| M. marinum M | MMAR_5412 | gltD | 5e-40 | 30.30% (439) | NADH-dependent glutamate synthase (small subunit) GltD |
| M. marinum M | MMAR_5477 | trxB2 | 2e-13 | 28.12% (320) | thioredoxin reductase TrxB2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3888c | gltD | 2e-40 | 30.59% (438) | glutamate synthase subunit beta |
| M. gilvum PYR-GCK | Mflv_3592 | - | 1e-42 | 31.46% (445) | glutamate synthase, NADH/NADPH, small subunit |
| M. tuberculosis H37Rv | Rv3858c | gltD | 2e-40 | 30.59% (438) | glutamate synthase subunit beta |
| M. leprae Br4923 | MLBr_00062 | gltD | 3e-40 | 29.82% (436) | glutamate synthase subunit beta |
| M. abscessus ATCC 19977 | MAB_0092 | gltD | 5e-41 | 29.68% (438) | glutamate synthase subunit beta |
| M. avium 104 | MAV_0167 | gltD | 2e-42 | 30.39% (441) | glutamate synthase subunit beta |
| M. smegmatis MC2 155 | MSMEG_3226 | - | 3e-39 | 29.64% (496) | glutamate synthase, NADH/NADPH, small subunit |
| M. thermoresistible (build 8) | TH_2103 | gltD | 6e-39 | 30.93% (443) | PROBABLE NADH-DEPENDENT GLUTAMATE SYNTHASE (SMALL SUBUNIT) |
| M. ulcerans Agy99 | MUL_5036 | gltD | 3e-40 | 30.30% (439) | glutamate synthase subunit beta |
| M. vanbaalenii PYR-1 | Mvan_4108 | - | 0.0 | 77.82% (541) | putative glutamate synthase (NADPH) small subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00062|M.leprae_Br4923 MADPAGFLKYTQRELPRRRPVPLRLRDWKELYEEFD----KETLRNQASR
MAV_0167|M.avium_104 MADPSGFLKYTHRELPKRRPVPLRLRDWREVYEEFD----HGTLREQATR
Mb3888c|M.bovis_AF2122/97 MADPGGFLKYTHRKLPKRRPVPLRLRDWREVYEEFD----NESLRQQATR
Rv3858c|M.tuberculosis_H37Rv MADPGGFLKYTHRKLPKRRPVPLRLRDWREVYEEFD----NESLRQQATR
MUL_5036|M.ulcerans_Agy99 MADPTGFLKYTHRELPTRRPVPLRLRDWNEVYEDFD----NETLREQATR
TH_2103|M.thermoresistible__bu MADPRGFLKITSRETPQRRPVPLRLRDWKEVYEDFS----DDVLRRQASR
MAB_0092|M.abscessus_ATCC_1997 MGDPSGFLNHTTRELPKRRPVPLRLLDWKEVYEDFE----SSRLQIQASR
Mflv_3592|M.gilvum_PYR-GCK MADPHGFLEVAKVDAAKR-PVDERIGDWREVYETQDPHQRAGEVSQQARR
MSMEG_3226|M.smegmatis_MC2_155 MADPTGFLRVPKIEAAKR-PVDERVGDWREVYERQDPHERAGEVSQQARR
MMAR_3407|M.marinum_M MVPEQSPGDVPEVAAPAPEPAPEDVTPLPDLLHGRHR---AGPDRERRPV
Mvan_4108|M.vanbaalenii_PYR-1 ------------------MPGFDDVTPLANLAEGHSR---AGPVRDRRPV
* : :: . :
MLBr_00062|M.leprae_Br4923 CMDCGIPFCHN---GCPLGNLIPEWNDLVRRGRWHDAVERLHATNNFPDV
MAV_0167|M.avium_104 CMDCGIPFCHN---GCPLGNLIPEWNDLVRTGRWRDAIERLHATNNFPDF
Mb3888c|M.bovis_AF2122/97 CMDCGIPFCHN---GCPLGNLIPEWNDLVRRGRWRDAIERLHATNNFPDF
Rv3858c|M.tuberculosis_H37Rv CMDCGIPFCHN---GCPLGNLIPEWNDLVRRGRWRDAIERLHATNNFPDF
MUL_5036|M.ulcerans_Agy99 CMDCGIPFCHN---GCPLGNLIPEWNDLVRQGRWRDAIERLHATNNFPDF
TH_2103|M.thermoresistible__bu CMDCGIPFCHN---GCPLGNLIPEWNDLVYRDRWRDAIERLHATNNFPEF
MAB_0092|M.abscessus_ATCC_1997 CMDCGIPFCHK---GCPLGNLIPEWNDMVYRGNWREGIERLHATNNFPEF
Mflv_3592|M.gilvum_PYR-GCK CMDCGIPFCHSGSAGCPLGNLIPEWNDLVRRGRWDAASDRLHATNNFPEF
MSMEG_3226|M.smegmatis_MC2_155 CMDCGIPFCHSGTAGCPLGNLIPEWNDLVRRGRWDAASDRLHATNNFPEF
MMAR_3407|M.marinum_M YVDLLPPCNAG----CPAGENIQAWLAHARVGENERAWRQLTADNPFAAI
Mvan_4108|M.vanbaalenii_PYR-1 YVDLLPPCNAG----CPAGENIQAWLAHTTAGRHEAAWRQLTADNPFPAI
:* * ** *: * * . .. . :* * * *. .
MLBr_00062|M.leprae_Br4923 TGRLCPAPCESACVLGINQD----PVTIKQIELEIIDKAFDEGWVQPLPP
MAV_0167|M.avium_104 TGRLCPAPCEPACVLGINQD----PVTIKQIELEIIDNAFDEGWVQPLPP
Mb3888c|M.bovis_AF2122/97 TGRLCPAPCEPACVLGINQD----PVTIKQIELEIIDKAFDEGWVQPRPP
Rv3858c|M.tuberculosis_H37Rv TGRLCPAPCEPACVLGINQD----PVTIKQIELEIIDKAFDEGWVQPRPP
MUL_5036|M.ulcerans_Agy99 TGRLCPAPCEPACVLGINQD----PVTIKQIELEIIDNAFDEGWVKPLPP
TH_2103|M.thermoresistible__bu TGRLCPAPCEASCVLGINQD----AVTIKQVEVEIIDKAFDEGWVVPLPP
MAB_0092|M.abscessus_ATCC_1997 TGRLCPAPCEASCVLGINQD----PVTIKQVEVELIDNAFENDWVKPIPP
Mflv_3592|M.gilvum_PYR-GCK TGRLCPAPCEAACVLSISEEQTGGSVTIKRIEQTIADQAWMDGLVEPQPP
MSMEG_3226|M.smegmatis_MC2_155 TGRLCPAPCEAACVLSISEEHTGGSVTIKRIEQSIADQAWMDGTVEPQPA
MMAR_3407|M.marinum_M HGRVCYHPCESVCNRADLDA----SVSIHAVERFLGDTAREKQWRFQ-TA
Mvan_4108|M.vanbaalenii_PYR-1 HGRVCYHPCETVCNRGRLDA----AVSIHAVERFLGDLARERGWMFE-PP
**:* ***. * . : .*:*: :* : * * ..
MLBr_00062|M.leprae_Br4923 QQLTGKTVAVVGSGPAGLAAAQQLTRAGHRVTVFERDDRIGGLLRYGIPE
MAV_0167|M.avium_104 DKLTGKKVAVVGSGPAGLAAAQQLTRAGHSVTVFERSDKIGGLLRYGIPE
Mb3888c|M.bovis_AF2122/97 RKLTGQTVAVVGSGPAGLAAAQQLTRAGHTVTVFEREDRIGGLLRYGIPE
Rv3858c|M.tuberculosis_H37Rv RKLTGQTVAVVGSGPAGLAAAQQLTRAGHTVTVFEREDRIGGLLRYGIPE
MUL_5036|M.ulcerans_Agy99 QTLTGKTVAVVGSGPAGLAAAQQLTRAGHKVTVFEREDRIGGLLRYGIPE
TH_2103|M.thermoresistible__bu KKLTGKKVAVVGSGPAGLAAAQQLTRAGHAVTVFERADRIGGLLRYGIPE
MAB_0092|M.abscessus_ATCC_1997 EVKTGKKVAVVGSGPAGLAAAQQLTRAGHDVTVYERADRIGGLLRYGIPE
Mflv_3592|M.gilvum_PYR-GCK AIATGRKVAVVGSGPAGLAAAQQLTRAGHDVTVYERDDRIGGLMRYGIPE
MSMEG_3226|M.smegmatis_MC2_155 QISTGKSVAVVGSGPAGLAAAQQLTRAGHEVTVYERDDRLGGLLRYGIPE
MMAR_3407|M.marinum_M PQPTGKRVLVVGAGPSGLSAAYHLARLGHHVEVRDAGAEPGGMMRYGIPS
Mvan_4108|M.vanbaalenii_PYR-1 STRTGKRVLVVGAGPSGLSAAYHLARLGHHVEVRDAGAGPGGMMRYGIPN
**: * ***:**:**:** :*:* ** * * : **::*****.
MLBr_00062|M.leprae_Br4923 FKMEKRFLERRLDQLRAEGTEFRSGVNVGVDITADQLCADFDAVVLAGGA
MAV_0167|M.avium_104 FKMEKRVLDRRLDQMRAEGTEFRAGVNVGVDITADQLRADFDAVVLAGGA
Mb3888c|M.bovis_AF2122/97 FKMEKRHLDRRLDQMRSEGTEFRPGVNVGVDISAEKLRADFDAVVLAGGA
Rv3858c|M.tuberculosis_H37Rv FKMEKRHLDRRLDQMRSEGTEFRPGVNVGVDISAEKLRADFDAVVLAGGA
MUL_5036|M.ulcerans_Agy99 FKMEKRVLNRRLEQMQAEGTEFRAGVNVGVDITAEQLRADFDAVVLAGGA
TH_2103|M.thermoresistible__bu FKMEKRHIDRRLEQMTAEGTEFRTKVNVGVDLPVERLRAEFDAVVLAGGA
MAB_0092|M.abscessus_ATCC_1997 FKMEKRHIDRRLEQMRAEGTVFEAGVNVGVDITADHLRSNFDAVVLAGGA
Mflv_3592|M.gilvum_PYR-GCK YKLEKDTLNQRLAQMRAEGTRFVTECEVGVDLPIEQLRAQHDAVVLAVGA
MSMEG_3226|M.smegmatis_MC2_155 YKLEKATLNQRLAQMRAEGTRFVTECEVGVDVSVEQLRNRHQAVVLAVGA
MMAR_3407|M.marinum_M YRLPRDVLDAEIERIATLGVQLTTGHRV-EDLAEERETGNFDAVFVAVGA
Mvan_4108|M.vanbaalenii_PYR-1 YRLPRNVLDDELNRIAAMGVQLTCDHRV-DDLAAERDEGGFDAVFVAVGA
::: : :: .: :: : *. : .* *:. :: .:**.:* **
MLBr_00062|M.leprae_Br4923 ATWRELPIPGRELDGVHQAMEFLPWVNRVQEGD---DVLGDDGQPPITAK
MAV_0167|M.avium_104 TAWRDLPIPGRELDGIHQAMEFLPWGNRQASGDLDEADMDADGQPPITAK
Mb3888c|M.bovis_AF2122/97 TAWRELPIPGRELEGVHQAMEFLPWANRVQEGD---DVLDEDGQPPITAK
Rv3858c|M.tuberculosis_H37Rv TAWRELPIPGRELEGVHQAMEFLPWANRVQEGD---DVLDEDGQPPITAK
MUL_5036|M.ulcerans_Agy99 TAWRDLPIPGRELDGVHQAMEYLPWANRLQEGD---DVLGPDGQPPITAQ
TH_2103|M.thermoresistible__bu TAWRDLPIPGRELDGIHQAMEYLPWANRVQLGD---PVVDEHGEPPITAK
MAB_0092|M.abscessus_ATCC_1997 TAWRDLPVPGRELEGIYQAMEYLPWANKVQLGD---DVVDADGQPPITAK
Mflv_3592|M.gilvum_PYR-GCK LRARDNQVEGRDLDGVHLAMEHLVPANRECEGD---------GPSELSAA
MSMEG_3226|M.smegmatis_MC2_155 LRGRDNDVPGRELAGVHLAMEHLVPANRECEGD---------GPSPISAA
MMAR_3407|M.marinum_M HLAKRVNIPSRDARSLIDAVSFLHQVAAGEKPE----------------L
Mvan_4108|M.vanbaalenii_PYR-1 HLAKRVDIPVRDAAPMMDAVSFLRSVASGDAPA----------------I
: : *: : *:..*
MLBr_00062|M.leprae_Br4923 DKKVVIIGGGDTGADCLGAVHRQGAIHVHQLEIMPRPPDARAAS-TPWPT
MAV_0167|M.avium_104 GKKVIIIGGGDTGADCLGTVHRQGATEVHQFEIMPRPPDTRAPS-TPWPT
Mb3888c|M.bovis_AF2122/97 GKKVVIIGGGDTGADCLGTVHRQGAIAVHQFEIMPRPPDARAES-TPWPT
Rv3858c|M.tuberculosis_H37Rv GKKVVIIGGGDTGADCLGTVHRQGAIAVHQFEIMPRPPDARAES-TPWPT
MUL_5036|M.ulcerans_Agy99 GKKVVIIGGGDTGADCLGTAHRQGAANIHQFEIMPRPPEARAAS-TPWPT
TH_2103|M.thermoresistible__bu GKKVVIIGGGDTGADCLGTAHRQGAVSVHQFEIMPRPPETRAPS-TPWPT
MAB_0092|M.abscessus_ATCC_1997 GKRVIIIGGGDTGADCLGTAHRQGAASVQQFEIMPKPPESRSER-DPWPT
Mflv_3592|M.gilvum_PYR-GCK GKHVVIIGGGDTGADCLGTAHRQGAASVTQLDYNPEPPESRDDTRSPWPT
MSMEG_3226|M.smegmatis_MC2_155 GKHVVIIGGGDTGADCLGTAHRQGAASVTQLDYNPEPPERRDDTLSPWPT
MMAR_3407|M.marinum_M GSHVAVYGGGNTAMDAARVARRLGAQDTVIIYRRTEAQMPAHEE------
Mvan_4108|M.vanbaalenii_PYR-1 GRRVAVYGGGNTAMDAARVARRLGAEDTVILYRRTRDQMPAHAE------
. :* : ***:*. *. ..:* ** : ..
MLBr_00062|M.leprae_Br4923 YPLMYRV--SPAHEEGGERVFSVNTEEFVGKN-GRVHALKAHEVTMQ---
MAV_0167|M.avium_104 YPLMYRV--SSAHEEGGERVFSVNTEEFVGEN-GRVTALKAHEVTME---
Mb3888c|M.bovis_AF2122/97 YPLMYRV--SAAHEEGGERVFSVNTEAFVGTD-GRVSALRAHEVTML---
Rv3858c|M.tuberculosis_H37Rv YPLMYRV--SAAHEEGGERVFSVNTEAFVGTD-GRVSALRAHEVTML---
MUL_5036|M.ulcerans_Agy99 YPLMYRV--SAAHEEGGERVFSVNTEKFIGDD-GRVRALKAHEVAMQ---
TH_2103|M.thermoresistible__bu YPLMFRV--SSAHEEGGERVFSVNTEEFVGRD-GRVTGLRAHEVTMV---
MAB_0092|M.abscessus_ATCC_1997 YPTLFRV--ASAHEEGGERIYAVNTERFLGED-GKLTGLRAHEVVFN---
Mflv_3592|M.gilvum_PYR-GCK WPIMLRTKLSPAHAEGGRRRYQVAVQRFLGDDRGNLRALEIAEVQVERDD
MSMEG_3226|M.smegmatis_MC2_155 WPLVLRT--SPAHAEGGARRFEVAVQRFVGDDNGHVRAVEIAEVRVTRDA
MMAR_3407|M.marinum_M -----ER--QDAQREGVQINWLRTITAFDDHE------IRVEQMELD---
Mvan_4108|M.vanbaalenii_PYR-1 -----EA--EDAEREGVRINWLRTITAFDGPD------IQVEVMELD---
. *. ** : * . : :. : .
MLBr_00062|M.leprae_Br4923 -DGRFVKVENSVFELEADLVLLAMGFVGPEKPGLLTDLGVKFTDRGNVAR
MAV_0167|M.avium_104 -DGKFVKVEGSEFELEADLVLLAMGFVGPEKEGLLTDLGVKFTERGNVAR
Mb3888c|M.bovis_AF2122/97 -DGKFVKVEGSDFELEADLVLLAMGFVGPERAGLLTDLGVKFTEHGNVAR
Rv3858c|M.tuberculosis_H37Rv -DGKFVKVEGSDFELEADLVLLAMGFVGPERAGLLTDLGVKFTERGNVAR
MUL_5036|M.ulcerans_Agy99 -DGKFVKVEGSDFELEADLVLLAMGFVGPEKPGLLTDLGVKLTDRGNVAR
TH_2103|M.thermoresistible__bu -NGRFEKVEGTDFELEADLVLLAMGFVGPEREGLLTDLGVELTDRGNVAR
MAB_0092|M.abscessus_ATCC_1997 -AGKFEKVEGSDFELEADIVFLAMGFVGPEKPGLLESLGVEFTERGNVAR
Mflv_3592|M.gilvum_PYR-GCK SGRRHIVPVGESLQIPCDLALLAIGFDGVEHMPLLDKLGLSLNRRGTLAC
MSMEG_3226|M.smegmatis_MC2_155 EGRRQITPVGEPLEIPCDLALLAIGFEGVEHMPLLDDLHIELNKRGAIPC
MMAR_3407|M.marinum_M -DTGRPRPTGRFETLAADTVIMALGQET-ESAFMRTLPGVEFDRDGSVRV
Mvan_4108|M.vanbaalenii_PYR-1 -DSGRPQPTGRFETLPADTLIMAIGQQT-ESAFMRTLPGVEFDADGSVLV
. : .* ::*:* * : :.: * :
MLBr_00062|M.leprae_Br4923 GADFDTSVPGVFVAGDMGRGQSLIVWAIAEGRAAAAAVDRFLMGATALPA
MAV_0167|M.avium_104 GADFDTSIPGVFVAGDMGRGQSLIVWAIAEGRAAAAAADRFLMGATALPA
Mb3888c|M.bovis_AF2122/97 GDDFDTSVPGVFVAGDMGRGQSLIVWAIAEGRAAAAAVDRYLMGSSALPA
Rv3858c|M.tuberculosis_H37Rv GDDFDTSVPGVFVAGDMGRGQSLIVWAIAEGRAAAAAVDRYLMGSSALPA
MUL_5036|M.ulcerans_Agy99 GDDFETSAPGVFVAGDMGRGQSLIVWAIAEGRAAAAGVDRYLMGSTALPA
TH_2103|M.thermoresistible__bu DKDFQTSVPGVFVAGDMGRGQSLIVWAIAEGRAAAAGVDRYLMRRTALPR
MAB_0092|M.abscessus_ATCC_1997 DDAYATSVDGVFVAGDMGRGQSLIVWAIAEGRSAAAAVDAYLTGETALPA
Mflv_3592|M.gilvum_PYR-GCK GSDWQTDAPGVFVCGDAHRGASLIVWAIAEGRSAAHAVDAYLMGESDLPA
MSMEG_3226|M.smegmatis_MC2_155 GSDWQTDAPGVFVCGDAHRGASLVVWAIAEGRSAAHAVDAYLMGESDLPS
MMAR_3407|M.marinum_M SPTLMTGCPGVFAGGDMVPCERTVTVGVGHGKKAAHYIDAWLRGDPTQRP
Mvan_4108|M.vanbaalenii_PYR-1 SASLMTGCPGVFAGGDMVPSERTVTVGVGHGKKAARHIDAWLRGASAGRP
. *. ***. ** :. .:..*: ** * :* .
MLBr_00062|M.leprae_Br4923 PVKPSSLPLQ----------------------------------------
MAV_0167|M.avium_104 PVKPTAVPLQ----------------------------------------
Mb3888c|M.bovis_AF2122/97 PVKPTAAPLQ----------------------------------------
Rv3858c|M.tuberculosis_H37Rv PVKPTAAPLQ----------------------------------------
MUL_5036|M.ulcerans_Agy99 PIKPTAAPLQ----------------------------------------
TH_2103|M.thermoresistible__bu PIEPTAAPQR----------------------------------------
MAB_0092|M.abscessus_ATCC_1997 PIKPTAAPQR----------------------------------------
Mflv_3592|M.gilvum_PYR-GCK PVRPGALPLAVV--------------------------------------
MSMEG_3226|M.smegmatis_MC2_155 PVRPGTLPLAVV--------------------------------------
MMAR_3407|M.marinum_M PKHPVADFDRLHLWYFGDARRRQQPELSPEDRISGFDEVMGGLSAEAATF
Mvan_4108|M.vanbaalenii_PYR-1 PKHPTATFDALHLWYFGDTAQRRQPELEATTRVQGFDEVVGGLSTDEAVY
* .* :
MLBr_00062|M.leprae_Br4923 --------------------------------------------------
MAV_0167|M.avium_104 --------------------------------------------------
Mb3888c|M.bovis_AF2122/97 --------------------------------------------------
Rv3858c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_5036|M.ulcerans_Agy99 --------------------------------------------------
TH_2103|M.thermoresistible__bu --------------------------------------------------
MAB_0092|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3592|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3226|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_3407|M.marinum_M EAGRCLSCGNCFECDGCLGACPEDAVIKLGRGHRYRFDYDRCTGCATCYD
Mvan_4108|M.vanbaalenii_PYR-1 ESGRCLSCGNCFECDGCLGACPEDAVIKLGVGQRYRFDYDKCTGCAVCAD
MLBr_00062|M.leprae_Br4923 --------------
MAV_0167|M.avium_104 --------------
Mb3888c|M.bovis_AF2122/97 --------------
Rv3858c|M.tuberculosis_H37Rv --------------
MUL_5036|M.ulcerans_Agy99 --------------
TH_2103|M.thermoresistible__bu --------------
MAB_0092|M.abscessus_ATCC_1997 --------------
Mflv_3592|M.gilvum_PYR-GCK --------------
MSMEG_3226|M.smegmatis_MC2_155 --------------
MMAR_3407|M.marinum_M QCPVHAIEMIAEPR
Mvan_4108|M.vanbaalenii_PYR-1 QCPVHAIEMFGEPT