For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADPHGFLEVAKVDAAKRPVDERIGDWREVYETQDPHQRAGEVSQQARRCMDCGIPFCHSGSAGCPLGNLI PEWNDLVRRGRWDAASDRLHATNNFPEFTGRLCPAPCEAACVLSISEEQTGGSVTIKRIEQTIADQAWMD GLVEPQPPAIATGRKVAVVGSGPAGLAAAQQLTRAGHDVTVYERDDRIGGLMRYGIPEYKLEKDTLNQRL AQMRAEGTRFVTECEVGVDLPIEQLRAQHDAVVLAVGALRARDNQVEGRDLDGVHLAMEHLVPANRECEG DGPSELSAAGKHVVIIGGGDTGADCLGTAHRQGAASVTQLDYNPEPPESRDDTRSPWPTWPIMLRTKLSP AHAEGGRRRYQVAVQRFLGDDRGNLRALEIAEVQVERDDSGRRHIVPVGESLQIPCDLALLAIGFDGVEH MPLLDKLGLSLNRRGTLACGSDWQTDAPGVFVCGDAHRGASLIVWAIAEGRSAAHAVDAYLMGESDLPAP VRPGALPLAVV*
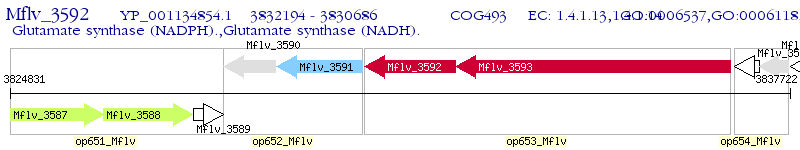
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3592 | - | - | 100% (502) | glutamate synthase, NADH/NADPH, small subunit |
| M. gilvum PYR-GCK | Mflv_1115 | gltD | e-151 | 56.50% (508) | glutamate synthase subunit beta |
| M. gilvum PYR-GCK | Mflv_1720 | - | 3e-10 | 32.45% (151) | 4Fe-4S ferredoxin iron-sulfur binding domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3888c | gltD | 1e-153 | 57.71% (506) | glutamate synthase subunit beta |
| M. tuberculosis H37Rv | Rv3858c | gltD | 1e-153 | 57.91% (506) | glutamate synthase subunit beta |
| M. leprae Br4923 | MLBr_00062 | gltD | 1e-150 | 57.00% (507) | glutamate synthase subunit beta |
| M. abscessus ATCC 19977 | MAB_0092 | gltD | 1e-149 | 56.24% (505) | glutamate synthase subunit beta |
| M. marinum M | MMAR_5412 | gltD | 1e-157 | 58.50% (506) | NADH-dependent glutamate synthase (small subunit) GltD |
| M. avium 104 | MAV_0167 | gltD | 1e-153 | 56.58% (509) | glutamate synthase subunit beta |
| M. smegmatis MC2 155 | MSMEG_3226 | - | 0.0 | 86.45% (502) | glutamate synthase, NADH/NADPH, small subunit |
| M. thermoresistible (build 8) | TH_2103 | gltD | 1e-151 | 57.43% (505) | PROBABLE NADH-DEPENDENT GLUTAMATE SYNTHASE (SMALL SUBUNIT) |
| M. ulcerans Agy99 | MUL_5036 | gltD | 1e-157 | 58.50% (506) | glutamate synthase subunit beta |
| M. vanbaalenii PYR-1 | Mvan_2823 | - | 0.0 | 91.83% (502) | glutamate synthases, NADH/NADPH, small subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00062|M.leprae_Br4923 MADPAGFLKYTQRELPRRRPVPLRLRDWKELYEEFD----KETLRNQASR
MAV_0167|M.avium_104 MADPSGFLKYTHRELPKRRPVPLRLRDWREVYEEFD----HGTLREQATR
Mb3888c|M.bovis_AF2122/97 MADPGGFLKYTHRKLPKRRPVPLRLRDWREVYEEFD----NESLRQQATR
Rv3858c|M.tuberculosis_H37Rv MADPGGFLKYTHRKLPKRRPVPLRLRDWREVYEEFD----NESLRQQATR
MMAR_5412|M.marinum_M MADPTGFLKYTHRELPTRRPVPLRLRDWNEVYEDFD----NETLREQATR
MUL_5036|M.ulcerans_Agy99 MADPTGFLKYTHRELPTRRPVPLRLRDWNEVYEDFD----NETLREQATR
MAB_0092|M.abscessus_ATCC_1997 MGDPSGFLNHTTRELPKRRPVPLRLLDWKEVYEDFE----SSRLQIQASR
TH_2103|M.thermoresistible__bu MADPRGFLKITSRETPQRRPVPLRLRDWKEVYEDFS----DDVLRRQASR
Mflv_3592|M.gilvum_PYR-GCK MADPHGFLEVAKVDAAKR-PVDERIGDWREVYETQDPHQRAGEVSQQARR
Mvan_2823|M.vanbaalenii_PYR-1 MADPHGFLEVAKVEAAKR-PVDERVGDWREVYERQDPHQRAGEVSQQARR
MSMEG_3226|M.smegmatis_MC2_155 MADPTGFLRVPKIEAAKR-PVDERVGDWREVYERQDPHERAGEVSQQARR
*.** ***. . . . * ** *: **.*:** . : ** *
MLBr_00062|M.leprae_Br4923 CMDCGIPFCHN---GCPLGNLIPEWNDLVRRGRWHDAVERLHATNNFPDV
MAV_0167|M.avium_104 CMDCGIPFCHN---GCPLGNLIPEWNDLVRTGRWRDAIERLHATNNFPDF
Mb3888c|M.bovis_AF2122/97 CMDCGIPFCHN---GCPLGNLIPEWNDLVRRGRWRDAIERLHATNNFPDF
Rv3858c|M.tuberculosis_H37Rv CMDCGIPFCHN---GCPLGNLIPEWNDLVRRGRWRDAIERLHATNNFPDF
MMAR_5412|M.marinum_M CMDCGIPFCHN---GCPLGNLIPEWNDLVRQGRWRDAIERLHATNNFPDF
MUL_5036|M.ulcerans_Agy99 CMDCGIPFCHN---GCPLGNLIPEWNDLVRQGRWRDAIERLHATNNFPDF
MAB_0092|M.abscessus_ATCC_1997 CMDCGIPFCHK---GCPLGNLIPEWNDMVYRGNWREGIERLHATNNFPEF
TH_2103|M.thermoresistible__bu CMDCGIPFCHN---GCPLGNLIPEWNDLVYRDRWRDAIERLHATNNFPEF
Mflv_3592|M.gilvum_PYR-GCK CMDCGIPFCHSGSAGCPLGNLIPEWNDLVRRGRWDAASDRLHATNNFPEF
Mvan_2823|M.vanbaalenii_PYR-1 CMDCGIPFCHSGTAGCPLGNLIPEWNDLVRRGRWDAASERLHATNNFPEF
MSMEG_3226|M.smegmatis_MC2_155 CMDCGIPFCHSGTAGCPLGNLIPEWNDLVRRGRWDAASDRLHATNNFPEF
**********. *************:* ..* . :*********:.
MLBr_00062|M.leprae_Br4923 TGRLCPAPCESACVLGINQD----PVTIKQIELEIIDKAFDEGWVQPLPP
MAV_0167|M.avium_104 TGRLCPAPCEPACVLGINQD----PVTIKQIELEIIDNAFDEGWVQPLPP
Mb3888c|M.bovis_AF2122/97 TGRLCPAPCEPACVLGINQD----PVTIKQIELEIIDKAFDEGWVQPRPP
Rv3858c|M.tuberculosis_H37Rv TGRLCPAPCEPACVLGINQD----PVTIKQIELEIIDKAFDEGWVQPRPP
MMAR_5412|M.marinum_M TGRLCPAPCEPACVLGINQD----PVTIKQIELEIIDNAFDEGWVKPLPP
MUL_5036|M.ulcerans_Agy99 TGRLCPAPCEPACVLGINQD----PVTIKQIELEIIDNAFDEGWVKPLPP
MAB_0092|M.abscessus_ATCC_1997 TGRLCPAPCEASCVLGINQD----PVTIKQVEVELIDNAFENDWVKPIPP
TH_2103|M.thermoresistible__bu TGRLCPAPCEASCVLGINQD----AVTIKQVEVEIIDKAFDEGWVVPLPP
Mflv_3592|M.gilvum_PYR-GCK TGRLCPAPCEAACVLSISEEQTGGSVTIKRIEQTIADQAWMDGLVEPQPP
Mvan_2823|M.vanbaalenii_PYR-1 TGRLCPAPCEAACVLSIAEEQTGGSVTIKRIEQTIADQAWMDGIVEPQPA
MSMEG_3226|M.smegmatis_MC2_155 TGRLCPAPCEAACVLSISEEHTGGSVTIKRIEQSIADQAWMDGTVEPQPA
**********.:***.* :: .****::* : *:*: :. * * *.
MLBr_00062|M.leprae_Br4923 QQLTGKTVAVVGSGPAGLAAAQQLTRAGHRVTVFERDDRIGGLLRYGIPE
MAV_0167|M.avium_104 DKLTGKKVAVVGSGPAGLAAAQQLTRAGHSVTVFERSDKIGGLLRYGIPE
Mb3888c|M.bovis_AF2122/97 RKLTGQTVAVVGSGPAGLAAAQQLTRAGHTVTVFEREDRIGGLLRYGIPE
Rv3858c|M.tuberculosis_H37Rv RKLTGQTVAVVGSGPAGLAAAQQLTRAGHTVTVFEREDRIGGLLRYGIPE
MMAR_5412|M.marinum_M QTLTGKTVAVVGSGPAGLAAAQQLTRAGHKVTVFEREDRIGGLLRYGIPE
MUL_5036|M.ulcerans_Agy99 QTLTGKTVAVVGSGPAGLAAAQQLTRAGHKVTVFEREDRIGGLLRYGIPE
MAB_0092|M.abscessus_ATCC_1997 EVKTGKKVAVVGSGPAGLAAAQQLTRAGHDVTVYERADRIGGLLRYGIPE
TH_2103|M.thermoresistible__bu KKLTGKKVAVVGSGPAGLAAAQQLTRAGHAVTVFERADRIGGLLRYGIPE
Mflv_3592|M.gilvum_PYR-GCK AIATGRKVAVVGSGPAGLAAAQQLTRAGHDVTVYERDDRIGGLMRYGIPE
Mvan_2823|M.vanbaalenii_PYR-1 AISTGKKVAVVGSGPAGLAAAQQLTRAGHDVTVYERDDRIGGLMRYGIPE
MSMEG_3226|M.smegmatis_MC2_155 QISTGKSVAVVGSGPAGLAAAQQLTRAGHEVTVYERDDRLGGLLRYGIPE
**:.********************** ***:** *::***:******
MLBr_00062|M.leprae_Br4923 FKMEKRFLERRLDQLRAEGTEFRSGVNVGVDITADQLCADFDAVVLAGGA
MAV_0167|M.avium_104 FKMEKRVLDRRLDQMRAEGTEFRAGVNVGVDITADQLRADFDAVVLAGGA
Mb3888c|M.bovis_AF2122/97 FKMEKRHLDRRLDQMRSEGTEFRPGVNVGVDISAEKLRADFDAVVLAGGA
Rv3858c|M.tuberculosis_H37Rv FKMEKRHLDRRLDQMRSEGTEFRPGVNVGVDISAEKLRADFDAVVLAGGA
MMAR_5412|M.marinum_M FKMEKRVLNRRLEQMQAEGTEFRAGVNVGVDIRAEQLRADFDAVVLAGGA
MUL_5036|M.ulcerans_Agy99 FKMEKRVLNRRLEQMQAEGTEFRAGVNVGVDITAEQLRADFDAVVLAGGA
MAB_0092|M.abscessus_ATCC_1997 FKMEKRHIDRRLEQMRAEGTVFEAGVNVGVDITADHLRSNFDAVVLAGGA
TH_2103|M.thermoresistible__bu FKMEKRHIDRRLEQMTAEGTEFRTKVNVGVDLPVERLRAEFDAVVLAGGA
Mflv_3592|M.gilvum_PYR-GCK YKLEKDTLNQRLAQMRAEGTRFVTECEVGVDLPIEQLRAQHDAVVLAVGA
Mvan_2823|M.vanbaalenii_PYR-1 YKLEKSILNQRLAQMRAEGTRFVTECEVGVDLPVEQLRAQHDAVVLAVGA
MSMEG_3226|M.smegmatis_MC2_155 YKLEKATLNQRLAQMRAEGTRFVTECEVGVDVSVEQLRNRHQAVVLAVGA
:*:** :::** *: :*** * . :****: ::* .:***** **
MLBr_00062|M.leprae_Br4923 ATWRELPIPGRELDGVHQAMEFLPWVNRVQEGD---DVLGDDGQPPITAK
MAV_0167|M.avium_104 TAWRDLPIPGRELDGIHQAMEFLPWGNRQASGDLDEADMDADGQPPITAK
Mb3888c|M.bovis_AF2122/97 TAWRELPIPGRELEGVHQAMEFLPWANRVQEGD---DVLDEDGQPPITAK
Rv3858c|M.tuberculosis_H37Rv TAWRELPIPGRELEGVHQAMEFLPWANRVQEGD---DVLDEDGQPPITAK
MMAR_5412|M.marinum_M TAWRDLPIPGRELDGVHQAMEYLPWANRLQEGD---DVLGPDGQPPITAQ
MUL_5036|M.ulcerans_Agy99 TAWRDLPIPGRELDGVHQAMEYLPWANRLQEGD---DVLGPDGQPPITAQ
MAB_0092|M.abscessus_ATCC_1997 TAWRDLPVPGRELEGIYQAMEYLPWANKVQLGD---DVVDADGQPPITAK
TH_2103|M.thermoresistible__bu TAWRDLPIPGRELDGIHQAMEYLPWANRVQLGD---PVVDEHGEPPITAK
Mflv_3592|M.gilvum_PYR-GCK LRARDNQVEGRDLDGVHLAMEHLVPANRECEGD---------GPSELSAA
Mvan_2823|M.vanbaalenii_PYR-1 LRARDNDVEGRDLAGVHLAMEHLVPANKECEGD---------GPSGVSAE
MSMEG_3226|M.smegmatis_MC2_155 LRGRDNDVPGRELAGVHLAMEHLVPANRECEGD---------GPSPISAA
*: : **:* *:: ***.* *: ** * . ::*
MLBr_00062|M.leprae_Br4923 DKKVVIIGGGDTGADCLGAVHRQGAIHVHQLEIMPRPPDARAAS-TPWPT
MAV_0167|M.avium_104 GKKVIIIGGGDTGADCLGTVHRQGATEVHQFEIMPRPPDTRAPS-TPWPT
Mb3888c|M.bovis_AF2122/97 GKKVVIIGGGDTGADCLGTVHRQGAIAVHQFEIMPRPPDARAES-TPWPT
Rv3858c|M.tuberculosis_H37Rv GKKVVIIGGGDTGADCLGTVHRQGAIAVHQFEIMPRPPDARAES-TPWPT
MMAR_5412|M.marinum_M GKKVVIIGGGDTGADCLGTAHRQGAANIHQFEIMPRPPEARAAS-TPWPT
MUL_5036|M.ulcerans_Agy99 GKKVVIIGGGDTGADCLGTAHRQGAANIHQFEIMPRPPEARAAS-TPWPT
MAB_0092|M.abscessus_ATCC_1997 GKRVIIIGGGDTGADCLGTAHRQGAASVQQFEIMPKPPESRSER-DPWPT
TH_2103|M.thermoresistible__bu GKKVVIIGGGDTGADCLGTAHRQGAVSVHQFEIMPRPPETRAPS-TPWPT
Mflv_3592|M.gilvum_PYR-GCK GKHVVIIGGGDTGADCLGTAHRQGAASVTQLDYNPEPPESRDDTRSPWPT
Mvan_2823|M.vanbaalenii_PYR-1 GKHVVIIGGGDTGADCLGTAHRQGATSVTQLDYNPEPPELRDESRSPWPT
MSMEG_3226|M.smegmatis_MC2_155 GKHVVIIGGGDTGADCLGTAHRQGAASVTQLDYNPEPPERRDDTLSPWPT
.*:*:*************:.***** : *:: *.**: * ****
MLBr_00062|M.leprae_Br4923 YPLMYRV--SPAHEEGGERVFSVNTEEFVGKN-GRVHALKAHEVTMQ---
MAV_0167|M.avium_104 YPLMYRV--SSAHEEGGERVFSVNTEEFVGEN-GRVTALKAHEVTME---
Mb3888c|M.bovis_AF2122/97 YPLMYRV--SAAHEEGGERVFSVNTEAFVGTD-GRVSALRAHEVTML---
Rv3858c|M.tuberculosis_H37Rv YPLMYRV--SAAHEEGGERVFSVNTEAFVGTD-GRVSALRAHEVTML---
MMAR_5412|M.marinum_M YPLMYRV--SAAHEEGGERVFSVNTEKFIGDD-GRVRALKAHEVAMQ---
MUL_5036|M.ulcerans_Agy99 YPLMYRV--SAAHEEGGERVFSVNTEKFIGDD-GRVRALKAHEVAMQ---
MAB_0092|M.abscessus_ATCC_1997 YPTLFRV--ASAHEEGGERIYAVNTERFLGED-GKLTGLRAHEVVFN---
TH_2103|M.thermoresistible__bu YPLMFRV--SSAHEEGGERVFSVNTEEFVGRD-GRVTGLRAHEVTMV---
Mflv_3592|M.gilvum_PYR-GCK WPIMLRTKLSPAHAEGGRRRYQVAVQRFLGDDRGNLRALEIAEVQVERDD
Mvan_2823|M.vanbaalenii_PYR-1 WPIVLRTRLSPAHAEGGHRRYEVAVQRFLGDDRGNLRAMEIAEVKVERDE
MSMEG_3226|M.smegmatis_MC2_155 WPLVLRT--SPAHAEGGARRFEVAVQRFVGDDNGHVRAVEIAEVRVTRDA
:* : *. :.** *** * : * .: *:* : *.: .:. ** .
MLBr_00062|M.leprae_Br4923 -DGRFVKVENSVFELEADLVLLAMGFVGPEKPGLLTDLGVKFTDRGNVAR
MAV_0167|M.avium_104 -DGKFVKVEGSEFELEADLVLLAMGFVGPEKEGLLTDLGVKFTERGNVAR
Mb3888c|M.bovis_AF2122/97 -DGKFVKVEGSDFELEADLVLLAMGFVGPERAGLLTDLGVKFTEHGNVAR
Rv3858c|M.tuberculosis_H37Rv -DGKFVKVEGSDFELEADLVLLAMGFVGPERAGLLTDLGVKFTERGNVAR
MMAR_5412|M.marinum_M -DGKFVKVEGSDFELEADLVLLAMGFVGPEKPGLLTDLGVKLTDRGNVAR
MUL_5036|M.ulcerans_Agy99 -DGKFVKVEGSDFELEADLVLLAMGFVGPEKPGLLTDLGVKLTDRGNVAR
MAB_0092|M.abscessus_ATCC_1997 -AGKFEKVEGSDFELEADIVFLAMGFVGPEKPGLLESLGVEFTERGNVAR
TH_2103|M.thermoresistible__bu -NGRFEKVEGTDFELEADLVLLAMGFVGPEREGLLTDLGVELTDRGNVAR
Mflv_3592|M.gilvum_PYR-GCK SGRRHIVPVGESLQIPCDLALLAIGFDGVEHMPLLDKLGLSLNRRGTLAC
Mvan_2823|M.vanbaalenii_PYR-1 AGRRHIVPVGESLEIPCELALLAIGFEGVEHMALLDKLGLTLSRRGTVPC
MSMEG_3226|M.smegmatis_MC2_155 EGRRQITPVGEPLEIPCDLALLAIGFEGVEHMPLLDDLHIELNKRGAIPC
: . ::: .::.:**:** * *: ** .* : :. :* :.
MLBr_00062|M.leprae_Br4923 GADFDTSVPGVFVAGDMGRGQSLIVWAIAEGRAAAAAVDRFLMGATALPA
MAV_0167|M.avium_104 GADFDTSIPGVFVAGDMGRGQSLIVWAIAEGRAAAAAADRFLMGATALPA
Mb3888c|M.bovis_AF2122/97 GDDFDTSVPGVFVAGDMGRGQSLIVWAIAEGRAAAAAVDRYLMGSSALPA
Rv3858c|M.tuberculosis_H37Rv GDDFDTSVPGVFVAGDMGRGQSLIVWAIAEGRAAAAAVDRYLMGSSALPA
MMAR_5412|M.marinum_M GDDFETSAPGVFVAGDMGRGQSLIVWAIAEGRAAAAGVDRYLMGSTALPA
MUL_5036|M.ulcerans_Agy99 GDDFETSAPGVFVAGDMGRGQSLIVWAIAEGRAAAAGVDRYLMGSTALPA
MAB_0092|M.abscessus_ATCC_1997 DDAYATSVDGVFVAGDMGRGQSLIVWAIAEGRSAAAAVDAYLTGETALPA
TH_2103|M.thermoresistible__bu DKDFQTSVPGVFVAGDMGRGQSLIVWAIAEGRAAAAGVDRYLMRRTALPR
Mflv_3592|M.gilvum_PYR-GCK GSDWQTDAPGVFVCGDAHRGASLIVWAIAEGRSAAHAVDAYLMGESDLPA
Mvan_2823|M.vanbaalenii_PYR-1 GSDWQTDAPGVFVCGDAHRGASLIVWAIAEGRSAAHAVDAYLMGDSDLPA
MSMEG_3226|M.smegmatis_MC2_155 GSDWQTDAPGVFVCGDAHRGASLVVWAIAEGRSAAHAVDAYLMGESDLPS
. : *. ****.** ** **:********:** ..* :* : **
MLBr_00062|M.leprae_Br4923 PVKPSSLPLQ--
MAV_0167|M.avium_104 PVKPTAVPLQ--
Mb3888c|M.bovis_AF2122/97 PVKPTAAPLQ--
Rv3858c|M.tuberculosis_H37Rv PVKPTAAPLQ--
MMAR_5412|M.marinum_M PIKPTAAPLQ--
MUL_5036|M.ulcerans_Agy99 PIKPTAAPLQ--
MAB_0092|M.abscessus_ATCC_1997 PIKPTAAPQR--
TH_2103|M.thermoresistible__bu PIEPTAAPQR--
Mflv_3592|M.gilvum_PYR-GCK PVRPGALPLAVV
Mvan_2823|M.vanbaalenii_PYR-1 PVRPGTLPLAVV
MSMEG_3226|M.smegmatis_MC2_155 PVRPGTLPLAVV
*:.* : *