For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MADPAGFLKYTQRELPRRRPVPLRLRDWKELYEEFDKETLRNQASRCMDCGIPFCHNGCPLGNLIPEWND LVRRGRWHDAVERLHATNNFPDVTGRLCPAPCESACVLGINQDPVTIKQIELEIIDKAFDEGWVQPLPPQ QLTGKTVAVVGSGPAGLAAAQQLTRAGHRVTVFERDDRIGGLLRYGIPEFKMEKRFLERRLDQLRAEGTE FRSGVNVGVDITADQLCADFDAVVLAGGAATWRELPIPGRELDGVHQAMEFLPWVNRVQEGDDVLGDDGQ PPITAKDKKVVIIGGGDTGADCLGAVHRQGAIHVHQLEIMPRPPDARAASTPWPTYPLMYRVSPAHEEGG ERVFSVNTEEFVGKNGRVHALKAHEVTMQDGRFVKVENSVFELEADLVLLAMGFVGPEKPGLLTDLGVKF TDRGNVARGADFDTSVPGVFVAGDMGRGQSLIVWAIAEGRAAAAAVDRFLMGATALPAPVKPSSLPLQ
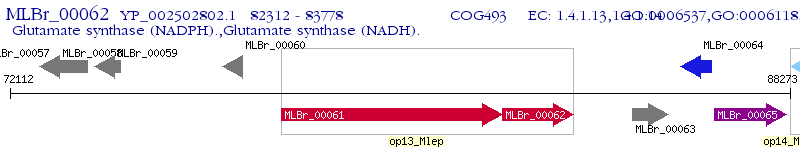
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00062 | gltD | - | 100% (488) | glutamate synthase subunit beta |
| M. leprae Br4923 | MLBr_02134 | fprB | 5e-14 | 36.25% (160) | ferredoxin, ferredoxin-NADP reductase |
| M. leprae Br4923 | MLBr_00666 | fprA | 3e-10 | 29.27% (164) | putative NADPH-ferredoxin reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3888c | gltD | 0.0 | 87.50% (488) | glutamate synthase subunit beta |
| M. gilvum PYR-GCK | Mflv_1115 | gltD | 0.0 | 78.53% (489) | glutamate synthase subunit beta |
| M. tuberculosis H37Rv | Rv3858c | gltD | 0.0 | 87.70% (488) | glutamate synthase subunit beta |
| M. abscessus ATCC 19977 | MAB_0092 | gltD | 0.0 | 72.84% (486) | glutamate synthase subunit beta |
| M. marinum M | MMAR_5412 | gltD | 0.0 | 86.68% (488) | NADH-dependent glutamate synthase (small subunit) GltD |
| M. avium 104 | MAV_0167 | gltD | 0.0 | 87.17% (491) | glutamate synthase subunit beta |
| M. smegmatis MC2 155 | MSMEG_6458 | gltD | 0.0 | 78.60% (486) | glutamate synthase subunit beta |
| M. thermoresistible (build 8) | TH_2103 | gltD | 0.0 | 80.25% (486) | PROBABLE NADH-DEPENDENT GLUTAMATE SYNTHASE (SMALL SUBUNIT) |
| M. ulcerans Agy99 | MUL_5036 | gltD | 0.0 | 86.89% (488) | glutamate synthase subunit beta |
| M. vanbaalenii PYR-1 | Mvan_5697 | gltD | 0.0 | 79.63% (486) | glutamate synthase subunit beta |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1115|M.gilvum_PYR-GCK MADPRGFLKYTHRETAKRRPVDLRLKDWKEVYEDFSHDTLKVQATRCMDC
Mvan_5697|M.vanbaalenii_PYR-1 MADPRGFLKHTHRETPKRRPVDLRLKDWKEVYEDFSHDTLQVQASRCMDC
MLBr_00062|M.leprae_Br4923 MADPAGFLKYTQRELPRRRPVPLRLRDWKELYEEFDKETLRNQASRCMDC
MAV_0167|M.avium_104 MADPSGFLKYTHRELPKRRPVPLRLRDWREVYEEFDHGTLREQATRCMDC
Mb3888c|M.bovis_AF2122/97 MADPGGFLKYTHRKLPKRRPVPLRLRDWREVYEEFDNESLRQQATRCMDC
Rv3858c|M.tuberculosis_H37Rv MADPGGFLKYTHRKLPKRRPVPLRLRDWREVYEEFDNESLRQQATRCMDC
MMAR_5412|M.marinum_M MADPTGFLKYTHRELPTRRPVPLRLRDWNEVYEDFDNETLREQATRCMDC
MUL_5036|M.ulcerans_Agy99 MADPTGFLKYTHRELPTRRPVPLRLRDWNEVYEDFDNETLREQATRCMDC
TH_2103|M.thermoresistible__bu MADPRGFLKITSRETPQRRPVPLRLRDWKEVYEDFSDDVLRRQASRCMDC
MSMEG_6458|M.smegmatis_MC2_155 MADPRGFLKHTTRELPKRRPVDLRLKDWKEVYEDFDHAPLQTQASRCMDC
MAB_0092|M.abscessus_ATCC_1997 MGDPSGFLNHTTRELPKRRPVPLRLLDWKEVYEDFESSRLQIQASRCMDC
*.** ***: * *: . **** *** **.*:**:*. *: **:*****
Mflv_1115|M.gilvum_PYR-GCK GIPFCHNGCPLGNLIPEWNDLVRTGRWRDAIERLHATNNFPEFTGRLCPA
Mvan_5697|M.vanbaalenii_PYR-1 GIPFCHNGCPLGNLIPEWNDLVRTGRWHDAIERLHATNNFPEFTGRLCPA
MLBr_00062|M.leprae_Br4923 GIPFCHNGCPLGNLIPEWNDLVRRGRWHDAVERLHATNNFPDVTGRLCPA
MAV_0167|M.avium_104 GIPFCHNGCPLGNLIPEWNDLVRTGRWRDAIERLHATNNFPDFTGRLCPA
Mb3888c|M.bovis_AF2122/97 GIPFCHNGCPLGNLIPEWNDLVRRGRWRDAIERLHATNNFPDFTGRLCPA
Rv3858c|M.tuberculosis_H37Rv GIPFCHNGCPLGNLIPEWNDLVRRGRWRDAIERLHATNNFPDFTGRLCPA
MMAR_5412|M.marinum_M GIPFCHNGCPLGNLIPEWNDLVRQGRWRDAIERLHATNNFPDFTGRLCPA
MUL_5036|M.ulcerans_Agy99 GIPFCHNGCPLGNLIPEWNDLVRQGRWRDAIERLHATNNFPDFTGRLCPA
TH_2103|M.thermoresistible__bu GIPFCHNGCPLGNLIPEWNDLVYRDRWRDAIERLHATNNFPEFTGRLCPA
MSMEG_6458|M.smegmatis_MC2_155 GIPFCHNGCPLGNLIPEWNDLVYRDRWRDAIERLHATNNFPEFTGRLCPA
MAB_0092|M.abscessus_ATCC_1997 GIPFCHKGCPLGNLIPEWNDMVYRGNWREGIERLHATNNFPEFTGRLCPA
******:*************:* ..*::.:**********:.*******
Mflv_1115|M.gilvum_PYR-GCK PCEGSCVLGINQDPVTIKQVEVEIIDNAFDEGWVVPLPPTKLTGKTVAVV
Mvan_5697|M.vanbaalenii_PYR-1 PCEGSCVLGINQDPVTIKQVEVEIIDNAFAEGWVVPLPPTKLTGKTVAVV
MLBr_00062|M.leprae_Br4923 PCESACVLGINQDPVTIKQIELEIIDKAFDEGWVQPLPPQQLTGKTVAVV
MAV_0167|M.avium_104 PCEPACVLGINQDPVTIKQIELEIIDNAFDEGWVQPLPPDKLTGKKVAVV
Mb3888c|M.bovis_AF2122/97 PCEPACVLGINQDPVTIKQIELEIIDKAFDEGWVQPRPPRKLTGQTVAVV
Rv3858c|M.tuberculosis_H37Rv PCEPACVLGINQDPVTIKQIELEIIDKAFDEGWVQPRPPRKLTGQTVAVV
MMAR_5412|M.marinum_M PCEPACVLGINQDPVTIKQIELEIIDNAFDEGWVKPLPPQTLTGKTVAVV
MUL_5036|M.ulcerans_Agy99 PCEPACVLGINQDPVTIKQIELEIIDNAFDEGWVKPLPPQTLTGKTVAVV
TH_2103|M.thermoresistible__bu PCEASCVLGINQDAVTIKQVEVEIIDKAFDEGWVVPLPPKKLTGKKVAVV
MSMEG_6458|M.smegmatis_MC2_155 PCEAACVLGINQDPVTIKQVEVEIIDHAFDQGWVVPKMPDVQTGKKVAVV
MAB_0092|M.abscessus_ATCC_1997 PCEASCVLGINQDPVTIKQVEVELIDNAFENDWVKPIPPEVKTGKKVAVV
*** :********.*****:*:*:**:** :.** * * **:.****
Mflv_1115|M.gilvum_PYR-GCK GSGPAGLAAAQQLTRAGHRVTVFERDDRIGGLLRYGIPEFKMEKHHIDRR
Mvan_5697|M.vanbaalenii_PYR-1 GSGPAGLAAAQQLTRAGHKVTVFERDDRIGGLLRYGIPEFKMEKRHIDRR
MLBr_00062|M.leprae_Br4923 GSGPAGLAAAQQLTRAGHRVTVFERDDRIGGLLRYGIPEFKMEKRFLERR
MAV_0167|M.avium_104 GSGPAGLAAAQQLTRAGHSVTVFERSDKIGGLLRYGIPEFKMEKRVLDRR
Mb3888c|M.bovis_AF2122/97 GSGPAGLAAAQQLTRAGHTVTVFEREDRIGGLLRYGIPEFKMEKRHLDRR
Rv3858c|M.tuberculosis_H37Rv GSGPAGLAAAQQLTRAGHTVTVFEREDRIGGLLRYGIPEFKMEKRHLDRR
MMAR_5412|M.marinum_M GSGPAGLAAAQQLTRAGHKVTVFEREDRIGGLLRYGIPEFKMEKRVLNRR
MUL_5036|M.ulcerans_Agy99 GSGPAGLAAAQQLTRAGHKVTVFEREDRIGGLLRYGIPEFKMEKRVLNRR
TH_2103|M.thermoresistible__bu GSGPAGLAAAQQLTRAGHAVTVFERADRIGGLLRYGIPEFKMEKRHIDRR
MSMEG_6458|M.smegmatis_MC2_155 GSGPAGLAAAQQLTRAGHTVTVFERADRIGGLLRYGIPEFKMEKRHIDRR
MAB_0092|M.abscessus_ATCC_1997 GSGPAGLAAAQQLTRAGHDVTVYERADRIGGLLRYGIPEFKMEKRHIDRR
****************** ***:** *:****************: ::**
Mflv_1115|M.gilvum_PYR-GCK LDQMRAEGTEFRTGVNVGKDITAAQLRQEFDAVVLAGGATARRDLPIPGR
Mvan_5697|M.vanbaalenii_PYR-1 LEQMRAEGTEFRAGVNVGVDITAAQLRSDFDAVVLAGGATARRDLPIPGR
MLBr_00062|M.leprae_Br4923 LDQLRAEGTEFRSGVNVGVDITADQLCADFDAVVLAGGAATWRELPIPGR
MAV_0167|M.avium_104 LDQMRAEGTEFRAGVNVGVDITADQLRADFDAVVLAGGATAWRDLPIPGR
Mb3888c|M.bovis_AF2122/97 LDQMRSEGTEFRPGVNVGVDISAEKLRADFDAVVLAGGATAWRELPIPGR
Rv3858c|M.tuberculosis_H37Rv LDQMRSEGTEFRPGVNVGVDISAEKLRADFDAVVLAGGATAWRELPIPGR
MMAR_5412|M.marinum_M LEQMQAEGTEFRAGVNVGVDIRAEQLRADFDAVVLAGGATAWRDLPIPGR
MUL_5036|M.ulcerans_Agy99 LEQMQAEGTEFRAGVNVGVDITAEQLRADFDAVVLAGGATAWRDLPIPGR
TH_2103|M.thermoresistible__bu LEQMTAEGTEFRTKVNVGVDLPVERLRAEFDAVVLAGGATAWRDLPIPGR
MSMEG_6458|M.smegmatis_MC2_155 LEQMAAEGTQFRPNVNVGVDITVAQLRLNYDAVVLAGGATAWRDLPIPGR
MAB_0092|M.abscessus_ATCC_1997 LEQMRAEGTVFEAGVNVGVDITADHLRSNFDAVVLAGGATAWRDLPVPGR
*:*: :*** *.. **** *: . :* ::*********:: *:**:***
Mflv_1115|M.gilvum_PYR-GCK ELDGIHQAMEFLPWANRFATGDLKDSVLDEDGEPPITAKGKKVIIIGGGD
Mvan_5697|M.vanbaalenii_PYR-1 ELDGIHQAMEYLPWANRVQLG---DPVVDADGQPPITAKGKKVIIIGGGD
MLBr_00062|M.leprae_Br4923 ELDGVHQAMEFLPWVNRVQEGD---DVLGDDGQPPITAKDKKVVIIGGGD
MAV_0167|M.avium_104 ELDGIHQAMEFLPWGNRQASGDLDEADMDADGQPPITAKGKKVIIIGGGD
Mb3888c|M.bovis_AF2122/97 ELEGVHQAMEFLPWANRVQEGD---DVLDEDGQPPITAKGKKVVIIGGGD
Rv3858c|M.tuberculosis_H37Rv ELEGVHQAMEFLPWANRVQEGD---DVLDEDGQPPITAKGKKVVIIGGGD
MMAR_5412|M.marinum_M ELDGVHQAMEYLPWANRLQEGD---DVLGPDGQPPITAQGKKVVIIGGGD
MUL_5036|M.ulcerans_Agy99 ELDGVHQAMEYLPWANRLQEGD---DVLGPDGQPPITAQGKKVVIIGGGD
TH_2103|M.thermoresistible__bu ELDGIHQAMEYLPWANRVQLGD---PVVDEHGEPPITAKGKKVVIIGGGD
MSMEG_6458|M.smegmatis_MC2_155 ELDGIHQAMEFLPWANRVQLGD---PVTDENGQPPITAKDKHVIIIGGGD
MAB_0092|M.abscessus_ATCC_1997 ELEGIYQAMEYLPWANKVQLGD---DVVDADGQPPITAKGKRVIIIGGGD
**:*::****:*** *: * . .*:*****:.*:*:******
Mflv_1115|M.gilvum_PYR-GCK TGADCLGASHRQGAVSVHQFEIMPRPPETRADSTPWPTYPLMFRVTSAHE
Mvan_5697|M.vanbaalenii_PYR-1 TGADCLGTSHRQGAVSVHQFEIMPRPPETRADSTPWPTYPLMYRVTSAHE
MLBr_00062|M.leprae_Br4923 TGADCLGAVHRQGAIHVHQLEIMPRPPDARAASTPWPTYPLMYRVSPAHE
MAV_0167|M.avium_104 TGADCLGTVHRQGATEVHQFEIMPRPPDTRAPSTPWPTYPLMYRVSSAHE
Mb3888c|M.bovis_AF2122/97 TGADCLGTVHRQGAIAVHQFEIMPRPPDARAESTPWPTYPLMYRVSAAHE
Rv3858c|M.tuberculosis_H37Rv TGADCLGTVHRQGAIAVHQFEIMPRPPDARAESTPWPTYPLMYRVSAAHE
MMAR_5412|M.marinum_M TGADCLGTAHRQGAANIHQFEIMPRPPEARAASTPWPTYPLMYRVSAAHE
MUL_5036|M.ulcerans_Agy99 TGADCLGTAHRQGAANIHQFEIMPRPPEARAASTPWPTYPLMYRVSAAHE
TH_2103|M.thermoresistible__bu TGADCLGTAHRQGAVSVHQFEIMPRPPETRAPSTPWPTYPLMFRVSSAHE
MSMEG_6458|M.smegmatis_MC2_155 TGADCLGTSHRQGAASIHQFEIMPRPPETRADSTPWPTYPLMFRVSSAHE
MAB_0092|M.abscessus_ATCC_1997 TGADCLGTAHRQGAASVQQFEIMPKPPESRSERDPWPTYPTLFRVASAHE
*******: ***** ::*:****:**::*: ****** ::**:.***
Mflv_1115|M.gilvum_PYR-GCK EGGERVYSVNTEAFVGSDGKVTGLRAHEVVMNAGKFEKVEGSDFEMDADL
Mvan_5697|M.vanbaalenii_PYR-1 EGGDRVYSVNTEQFVGEDGRVTGLRGHEVVMKAGKFEKVDGTDFEMDADL
MLBr_00062|M.leprae_Br4923 EGGERVFSVNTEEFVGKNGRVHALKAHEVTMQDGRFVKVENSVFELEADL
MAV_0167|M.avium_104 EGGERVFSVNTEEFVGENGRVTALKAHEVTMEDGKFVKVEGSEFELEADL
Mb3888c|M.bovis_AF2122/97 EGGERVFSVNTEAFVGTDGRVSALRAHEVTMLDGKFVKVEGSDFELEADL
Rv3858c|M.tuberculosis_H37Rv EGGERVFSVNTEAFVGTDGRVSALRAHEVTMLDGKFVKVEGSDFELEADL
MMAR_5412|M.marinum_M EGGERVFSVNTEKFIGDDGRVRALKAHEVAMQDGKFVKVEGSDFELEADL
MUL_5036|M.ulcerans_Agy99 EGGERVFSVNTEKFIGDDGRVRALKAHEVAMQDGKFVKVEGSDFELEADL
TH_2103|M.thermoresistible__bu EGGERVFSVNTEEFVGRDGRVTGLRAHEVTMVNGRFEKVEGTDFELEADL
MSMEG_6458|M.smegmatis_MC2_155 EGGERVFSVNTEEFIGENGHVTALRAHEVKMNAGKFEKVEGTDFELKADL
MAB_0092|M.abscessus_ATCC_1997 EGGERIYAVNTERFLGEDGKLTGLRAHEVVFNAGKFEKVEGSDFELEADI
***:*:::**** *:* :*:: .*:.*** : *:* **:.: **:.**:
Mflv_1115|M.gilvum_PYR-GCK VLLAMGFTGPEREGLLSDLGVEFNERGNVTRDDAFATSVPGVYVAGDMGR
Mvan_5697|M.vanbaalenii_PYR-1 VLLAMGFTGPEREGLLTDLGVEFNERGNVARDSAFATSVPGVFVAGDMGR
MLBr_00062|M.leprae_Br4923 VLLAMGFVGPEKPGLLTDLGVKFTDRGNVARGADFDTSVPGVFVAGDMGR
MAV_0167|M.avium_104 VLLAMGFVGPEKEGLLTDLGVKFTERGNVARGADFDTSIPGVFVAGDMGR
Mb3888c|M.bovis_AF2122/97 VLLAMGFVGPERAGLLTDLGVKFTEHGNVARGDDFDTSVPGVFVAGDMGR
Rv3858c|M.tuberculosis_H37Rv VLLAMGFVGPERAGLLTDLGVKFTERGNVARGDDFDTSVPGVFVAGDMGR
MMAR_5412|M.marinum_M VLLAMGFVGPEKPGLLTDLGVKLTDRGNVARGDDFETSAPGVFVAGDMGR
MUL_5036|M.ulcerans_Agy99 VLLAMGFVGPEKPGLLTDLGVKLTDRGNVARGDDFETSAPGVFVAGDMGR
TH_2103|M.thermoresistible__bu VLLAMGFVGPEREGLLTDLGVELTDRGNVARDKDFQTSVPGVFVAGDMGR
MSMEG_6458|M.smegmatis_MC2_155 VLLAMGFVGPEKEGLLDKLGVELTDRGNVKRDDNFQTSVPGVFVAGDMGR
MAB_0092|M.abscessus_ATCC_1997 VFLAMGFVGPEKPGLLESLGVEFTERGNVARDDAYATSVDGVFVAGDMGR
*:*****.***: *** .***::.::*** *. : ** **:*******
Mflv_1115|M.gilvum_PYR-GCK GQSLIVWAIAEGRAAAAAVDKYLMGNSALPAPIKPTAAPQR
Mvan_5697|M.vanbaalenii_PYR-1 GQSLIVWAIAEGRAAAAAADRYLVGHTALPAPIKPTAAPQR
MLBr_00062|M.leprae_Br4923 GQSLIVWAIAEGRAAAAAVDRFLMGATALPAPVKPSSLPLQ
MAV_0167|M.avium_104 GQSLIVWAIAEGRAAAAAADRFLMGATALPAPVKPTAVPLQ
Mb3888c|M.bovis_AF2122/97 GQSLIVWAIAEGRAAAAAVDRYLMGSSALPAPVKPTAAPLQ
Rv3858c|M.tuberculosis_H37Rv GQSLIVWAIAEGRAAAAAVDRYLMGSSALPAPVKPTAAPLQ
MMAR_5412|M.marinum_M GQSLIVWAIAEGRAAAAGVDRYLMGSTALPAPIKPTAAPLQ
MUL_5036|M.ulcerans_Agy99 GQSLIVWAIAEGRAAAAGVDRYLMGSTALPAPIKPTAAPLQ
TH_2103|M.thermoresistible__bu GQSLIVWAIAEGRAAAAGVDRYLMRRTALPRPIEPTAAPQR
MSMEG_6458|M.smegmatis_MC2_155 GQSLIVWAIAEGRAAAAGVDRYLMGRSALPAPIKPTAAPQR
MAB_0092|M.abscessus_ATCC_1997 GQSLIVWAIAEGRSAAAAVDAYLTGETALPAPIKPTAAPQR
*************:***..* :* :*** *::*:: * :