For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RALFARRSVQFGVGAVAVAAALTIAAAVLPPGTPGAAPSTDAANCPQVEVVFARGRFESPGPGVLGNAFI SSLSSKVGGRSMGSYAVKYPADTEVDIGANDMSGHIQYMVNSCPDTRLVLGGYSLGAAVTDVVLAVPMSA FGFENPLPPGTDQHIAAVALFGNGSQWVGPITNFSPLYNDRTIELCHGADPICSPADPNTWKQNWPQHLA SAYIQDGMVNEAADFVAGKL*
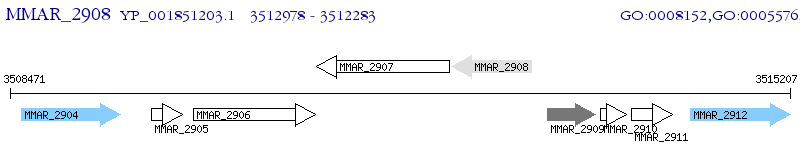
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2908 | - | - | 100% (231) | cutinase precursor |
| M. marinum M | MMAR_1097 | cut4 | 2e-44 | 44.87% (234) | cutinase precursor, Cut4 |
| M. marinum M | MMAR_3837 | - | 1e-40 | 43.04% (237) | cutinase precursor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2323 | cut2 | 7e-93 | 69.83% (232) | cutinase CUT2 |
| M. gilvum PYR-GCK | Mflv_4967 | - | 1e-79 | 65.89% (214) | cutinase |
| M. tuberculosis H37Rv | Rv2301 | cut2 | 8e-93 | 69.83% (232) | cutinase CUT2 |
| M. leprae Br4923 | MLBr_00099 | - | 2e-05 | 24.51% (257) | hypothetical protein MLBr_00099 |
| M. abscessus ATCC 19977 | MAB_3763 | - | 4e-48 | 42.53% (221) | cutinase cut2 precursor |
| M. avium 104 | MAV_2759 | - | 4e-88 | 83.61% (183) | serine esterase, cutinase family protein |
| M. smegmatis MC2 155 | MSMEG_1526 | - | 7e-88 | 69.72% (218) | cutinase |
| M. thermoresistible (build 8) | TH_3765 | - | 8e-85 | 65.33% (225) | PUTATIVE Cutinase |
| M. ulcerans Agy99 | MUL_0855 | cut4 | 5e-44 | 44.44% (234) | cutinase precursor, Cut4 |
| M. vanbaalenii PYR-1 | Mvan_1439 | - | 3e-79 | 66.05% (215) | cutinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4967|M.gilvum_PYR-GCK ---------------MIRQLRNLRVPYLVVALLFGAASLILP--------
Mvan_1439|M.vanbaalenii_PYR-1 ---------------MIRQLRKFPVLSALAALLSTAAALVAP--------
MMAR_2908|M.marinum_M MRALF-----------ARRSVQFGVGAVAVAAALTIAAAVLPPGTPGA--
MAV_2759|M.avium_104 --------------------------------------------------
Mb2323|M.bovis_AF2122/97 MNDLL-----------TRRLLTMGAAAAMLAAVLLLTPITVPAGYPGA--
Rv2301|M.tuberculosis_H37Rv MNDLL-----------TRRLLTMGAAAAMLAAVLLLTPITVPAGYPGA--
MSMEG_1526|M.smegmatis_MC2_155 MVEHS-----------SSPQLLKWLSAVVIVAATAVALVVAPTVTTRG--
TH_3765|M.thermoresistible__bu MVGTMGEVNSDLELPGTRRTVALLTAALFLAAGLVTGSAVSPASAP-L--
MAB_3763|M.abscessus_ATCC_1997 MTLQP-----------NPRPASKLLTAAARAAVVSVAAIMLTTGLAVV--
MUL_0855|M.ulcerans_Agy99 MWLFP-----LDAQKWARSGRGRTRLARRCAGLVATTIAACAVLLAFP--
MLBr_00099|M.leprae_Br4923 MVEKVPRKRHRVLAWTAALSMAAVVALAIVAVVILLRSAESPRSSLPPGV
Mflv_4967|M.gilvum_PYR-GCK --------------PAASAQACPQAELIFARGRIE------SPGAGVIGN
Mvan_1439|M.vanbaalenii_PYR-1 --------------PLASAQACPEVELIFARGRIE------SPGAGQIGN
MMAR_2908|M.marinum_M -------------APSTDAANCPQVEVVFARGRFE------SPGPGVLGN
MAV_2759|M.avium_104 ------------------------MQVVFARGRNE------SPGPGVLGN
Mb2323|M.bovis_AF2122/97 -------------VAPATAA-CPDAEVVFARGRFE------PPGIGTVGN
Rv2301|M.tuberculosis_H37Rv -------------VAPATAA-CPDAEVVFARGRFE------PPGIGTVGN
MSMEG_1526|M.smegmatis_MC2_155 -------------LPMAGAAPCADVEVIFARGRTE------PAGVGTLGN
TH_3765|M.thermoresistible__bu -------------APVAQAAPCPDVEVIFARGRVE------SPGPGIIGN
MAB_3763|M.abscessus_ATCC_1997 -------------IPATASAACPNVEVSFARGRSQ------PPGLGNIGD
MUL_0855|M.ulcerans_Agy99 -------------LVPKAAAACPDVQVVFARGTGE------PPGLGRVGN
MLBr_00099|M.leprae_Br4923 LPIPSTAPHPRKPRPAFQDVSCPDVQLLVVPGTWESSLQDNPLDPVQFPD
:: .. * : . . . :
Mflv_4967|M.gilvum_PYR-GCK ALAS----ALRNKTG-RDIDLYAVKYPADT--------------EVDIGA
Mvan_1439|M.vanbaalenii_PYR-1 ALVS----ALRSKTG-KSVDLYAVKYPADT--------------EVDLGA
MMAR_2908|M.marinum_M AFIS----SLSSKVGGRSMGSYAVKYPADT--------------EVDIGA
MAV_2759|M.avium_104 AFIS----ALRSRVN-RTVGVYAVQYPADT--------------EVAQGA
Mb2323|M.bovis_AF2122/97 AFVS----ALRSKVN-KNVGVYAVKYPADN--------------QIDVGA
Rv2301|M.tuberculosis_H37Rv AFVS----ALRSKVN-KNVGVYAVKYPADN--------------QIDVGA
MSMEG_1526|M.smegmatis_MC2_155 AFVN----ALRPKVK-KSMNVYAVKYPADT--------------EVDVGA
TH_3765|M.thermoresistible__bu AFVN----SLRSKTD-KNVSLYAVQYPADY--------------EIDIGA
MAB_3763|M.abscessus_ATCC_1997 AFVS----ALRNRVK--NLSVYAVRYDANT--------------DVGGGA
MUL_0855|M.ulcerans_Agy99 AMVA----SLRQKTD-QSIGAYAVNYPANKD-----------FLAAADGA
MLBr_00099|M.leprae_Br4923 ALLRNSTMTIGQQFPTSRVQTYTIPYTAQFHNPLSGDKQMTYNDSRAEGT
*: :: : : *:: * *: *:
Mflv_4967|M.gilvum_PYR-GCK NDMSQRIQYMARNCPDTRLVLGGYSLGAAVT----------------DIV
Mvan_1439|M.vanbaalenii_PYR-1 NDMSQRIQYMAGNCPDTRLVLGGYSLGAAVT----------------DVV
MMAR_2908|M.marinum_M NDMSGHIQYMVNSCPDTRLVLGGYSLGAAVT----------------DVV
MAV_2759|M.avium_104 NDMSRHIQDMVNSCPDTRLVLGGYSLGAAVT----------------DVV
Mb2323|M.bovis_AF2122/97 NDMSAHIQSMANSCPNTRLVPGGYSLGAAVT----------------DVV
Rv2301|M.tuberculosis_H37Rv NDMSAHIQSMANSCPNTRLVPGGYSLGAAVT----------------DVV
MSMEG_1526|M.smegmatis_MC2_155 NDMSNRIKYMMDNCPNTRLVIGGYSLGAAVA----------------DVV
TH_3765|M.thermoresistible__bu NDMSRRIQHMAATCPDTRLVLGGYSLGAAVT----------------DVV
MAB_3763|M.abscessus_ATCC_1997 NDLSAHLQQTANNCPDTKLVVGGYSLGAGVV----------------DTA
MUL_0855|M.ulcerans_Agy99 NDASDHVQQMTNECPNTKLVLGGYSQGAAVI----------------NIV
MLBr_00099|M.leprae_Br4923 RAMVQEMINVNNKCPLTSYVLVGFSQGAVIAGDITSDIGNGHGPVDDDLV
. .: ** * * *:* ** : : .
Mflv_4967|M.gilvum_PYR-GCK IAVP-----IAAFGFKNPLPD-----GVDRHIAAVALFGNGSQWVGPIAN
Mvan_1439|M.vanbaalenii_PYR-1 VAVP-----IAAFGFKSPLPD-----GVDRHIAAVALFGNGSQWVGPIAN
MMAR_2908|M.marinum_M LAVP-----MSAFGFENPLPP-----GTDQHIAAVALFGNGSQWVGPITN
MAV_2759|M.avium_104 LAAP-----IGAFGFDSPLPP-----GVDQHIAAVALFGNGSQWVGPITN
Mb2323|M.bovis_AF2122/97 LAVP-----TQMWGFTNPLPP-----GSDEHIAAVALFGNGSQWVGPITN
Rv2301|M.tuberculosis_H37Rv LAVP-----TQMWGFTNPLPP-----GSDEHIAAVALFGNGSQWVGPITN
MSMEG_1526|M.smegmatis_MC2_155 LAVP-----FTAFGFKNPLPA-----GADQHIAAVALFGNGAAWVGPITN
TH_3765|M.thermoresistible__bu LAVP-----ISALGFDSPLPA-----GVDRHIAAIALFGNGIAWAGPISN
MAB_3763|M.abscessus_ATCC_1997 LGVPGPVQIGNLFSFDNPVPP-----EVGDHIRAIVTFGNIVNRFAPVQS
MUL_0855|M.ulcerans_Agy99 TAAP-----LPGLGFTQPLPA-----QAAQHVAAVTLFGNPSGRAGGLMT
MLBr_00099|M.leprae_Br4923 LGVTLIADGRRQQGVGNDIGPNPPGEGAEVTLHEVPVLSGLGMTMTGARP
... .. . : : : :..
Mflv_4967|M.gilvum_PYR-GCK FTNPALRDRIVELCHGDDPICNPTAPENWQDNWPAHLASAYVGAGMVNQA
Mvan_1439|M.vanbaalenii_PYR-1 FTNPALRDRIIELCHGDDPICNPTAPENWQDYWPAHLAPAYIGAGMVNQA
MMAR_2908|M.marinum_M FS-PLYNDRTIELCHGADPICSPADPNTWKQNWPQHLASAYIQDGMVNEA
MAV_2759|M.avium_104 FN-PTYNDRTIELCHGADPICNPADPNTWKQNWPQHLAGAYIDAGMADQA
Mb2323|M.bovis_AF2122/97 FS-PAYNDRTIELCHGDDPVCHPADPNTWEANWPQHLAGAYVSSGMVNQA
Rv2301|M.tuberculosis_H37Rv FS-PAYNDRTIELCHGDDPVCHPADPNTWEANWPQHLAGAYVSSGMVNQA
MSMEG_1526|M.smegmatis_MC2_155 FS-PVYNDRTIELCHGADPICNPADPNTWQNNWPDHLAGAYISGGMVNQA
TH_3765|M.thermoresistible__bu FS-PLYAERTIELCHGADPICNPADPETWRDNWSDHAATKYVQSGMVDQA
MAB_3763|M.abscessus_ATCC_1997 LA-GAYGDRVLDLCNDGDPVCMDGNQDTWKS----HTS---YPPALINQA
MUL_0855|M.ulcerans_Agy99 ALSPHFGGKILNLCNTGDPICSDGDQ------WKAHTG---YVPGLTNKA
MLBr_00099|M.leprae_Br4923 GGFGVLHSRTNEICAPGDLICAAPAEAFSVANLPATLNTLASGAGQPIHA
: ::* * :* . .*
Mflv_4967|M.gilvum_PYR-GCK ADFIVGRL----------------------------
Mvan_1439|M.vanbaalenii_PYR-1 ADFIVGRL----------------------------
MMAR_2908|M.marinum_M ADFVAGKL----------------------------
MAV_2759|M.avium_104 ADFVAGKL----------------------------
Mb2323|M.bovis_AF2122/97 ADFVAGKLQ---------------------------
Rv2301|M.tuberculosis_H37Rv ADFVAGKLQ---------------------------
MSMEG_1526|M.smegmatis_MC2_155 ADFVASKL----------------------------
TH_3765|M.thermoresistible__bu ADFVAARL----------------------------
MAB_3763|M.abscessus_ATCC_1997 ADFAAARVN---------------------------
MUL_0855|M.ulcerans_Agy99 ASFVAGRI----------------------------
MLBr_00099|M.leprae_Br4923 MYATAQFWDLDGAPATVWTLNWVHRLIEGAPHPKHG
.