For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
YGALVTAADSTRTGLRECMQAAFRPRTGPPSASAILRSVLWPAAILSVLHRAIVLTTNGNITDDFKPVYR AVLNFRRGWDIYNEHFDYVDPHYLYPPGGTLLMAPFGYLPFAPSRYLFISINTAAILLAAYLLLRLFNFT LTSVAAPALVLAMFATETVTNTLVFTNINGCILLLEVLFLRWLLDGRVSHQWWAGIAIGFTLVLKPLLGP LLLLPLLNRQWRTLVTAIVIPIVINLAALPLINHPMDFFTRTLPYILGTRDYFNSSIQGNGIYFGLPTWL ILFLRLLFTAIAIASLWLLYRYYRTRDPLFWFTTSSGVLLLWSWLAMSLAQGYYSMMLFPFLMTITLRNS VIRNWPAWLGIYGFMTLDRWLLFNWMRWGRALEYLKITYGWSLLLIVTFTVLYFRYLDAKAEDRLDSGID PAWLTAERERASVDA*
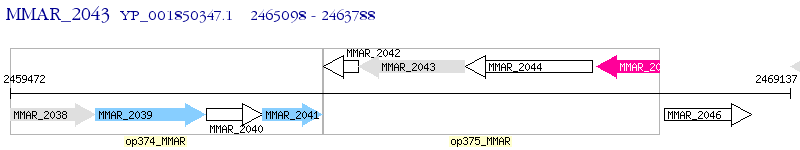
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2043 | - | - | 100% (436) | transmembrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2692 | - | 0.0 | 88.14% (430) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_3915 | - | 1e-173 | 70.22% (403) | putative integral membrane protein |
| M. tuberculosis H37Rv | Rv2673 | - | 0.0 | 88.14% (430) | integral membrane protein |
| M. leprae Br4923 | MLBr_01338 | - | 0.0 | 82.73% (440) | hypothetical protein MLBr_01338 |
| M. abscessus ATCC 19977 | MAB_2978 | - | 1e-157 | 63.32% (428) | hypothetical protein MAB_2978 |
| M. avium 104 | MAV_3565 | - | 0.0 | 82.56% (430) | hypothetical protein MAV_3565 |
| M. smegmatis MC2 155 | MSMEG_2785 | - | 1e-174 | 68.66% (434) | putative integral membrane protein |
| M. thermoresistible (build 8) | TH_0624 | - | 1e-164 | 70.48% (393) | POSSIBLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_3307 | - | 0.0 | 99.54% (436) | transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_2488 | - | 1e-175 | 69.51% (410) | putative integral membrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3915|M.gilvum_PYR-GCK -----------MLTAFR--------PRTAPPTTATVLRSILWPLAIMAVI
Mvan_2488|M.vanbaalenii_PYR-1 -------MRDLVLTAFR--------PRTSPPTTASVLRSILWPLAIMAVI
TH_0624|M.thermoresistible__bu -----------------------------------VLRSVLWPIAIMAVI
MAB_2978|M.abscessus_ATCC_1997 -----MVSVDALVTGFKN----SLAPKTAPPSVATILRSVLWPVAIMAVI
MSMEG_2785|M.smegmatis_MC2_155 MYCALVTATDSITTKLLN----AFRPRTSAPSTATVLRSVLWPIAILSVI
MMAR_2043|M.marinum_M MYGALVTAADSTRTGLRECMQAAFRPRTGPPSASAILRSVLWPAAILSVL
MUL_3307|M.ulcerans_Agy99 MYGALVTAADSTRTGLRECMQAAFRPRTGPPSASAILRSVLWPAAILSVL
Mb2692|M.bovis_AF2122/97 MYGALVTAADSIRTGLGASLLAGFRPRTGAPSTATILRSALWPAAVLSVL
Rv2673|M.tuberculosis_H37Rv MYGALVTAADSIRTGLGASLLAGFRPRTGAPSTATILRSALWPAAVLSVL
MLBr_01338|M.leprae_Br4923 MYGALVTAADSTQTGLRNWLLAVFHPRTHTPSTATIVRSALWPAAILSVL
MAV_3565|M.avium_104 MYGALVTAAES--TGLRDWIQAAFRPRTSAPSVATVLRSALWPIAIFSVL
::** *** *:::*:
Mflv_3915|M.gilvum_PYR-GCK HRSYVLVFNRYITDDFAPVYRAVINFKFGWDIYNEHFDHVDPHYLYPPGG
Mvan_2488|M.vanbaalenii_PYR-1 HRSYVLVFNRYITDDFAPVYRAVINFKFGWDIYNENFGHVDPHYLYPPGG
TH_0624|M.thermoresistible__bu HRSWVLSTNRSITDDFTPVYNAAVAFLRGQPVYTANFNHVDPHFLYPPGG
MAB_2978|M.abscessus_ATCC_1997 HRSYVLGTNGYITDDFGPVYRAVIAFKLHQPVYGENFNYVDPHYIYPPGG
MSMEG_2785|M.smegmatis_MC2_155 HRSYVLGTNGYITDDFGPVYRAVINFKLGLDIYNEQFDHVDPHYLYPPGG
MMAR_2043|M.marinum_M HRAIVLTTNGNITDDFKPVYRAVLNFRRGWDIYNEHFDYVDPHYLYPPGG
MUL_3307|M.ulcerans_Agy99 HRAIVLTTNGNITDDFKPVYRAVLNFRRGWDIYNEHFDYVDPHYLYPPGG
Mb2692|M.bovis_AF2122/97 HRSIVLTTNGNITDDFKPVYRAVLNFRRGWDIYNEHFDYVDPHYLYPPGG
Rv2673|M.tuberculosis_H37Rv HRSIVLTTNGNITDDFKPVYRAVLNFRRGWDIYNEHFDYVDPHYLYPPGG
MLBr_01338|M.leprae_Br4923 HRSTVITTNGNITDDFKPVYRAVLNFRHGWDIYNEHFDYVDPHYLYPPGG
MAV_3565|M.avium_104 HRSIVVTTNGNITDDFKPVYRAVLNFRRGWDIYNEHFDYVDPHYLYPPGG
**: *: * ***** ***.*.: * :* :*.:****::*****
Mflv_3915|M.gilvum_PYR-GCK TLIIAPFGYLPEVASRNWFIFFNTVAIILAGYFLLRLFNYTLSSVAAPAL
Mvan_2488|M.vanbaalenii_PYR-1 TLIIAPFGYLPEVASRNWFIFFNTVAIVLAGYFLLRLFNFTLASVAAPAL
TH_0624|M.thermoresistible__bu TLLMAPFGYLTPDVSRGWFIFFNTVAIVLAAYFLLRLFNFTLASVAAPAL
MAB_2978|M.abscessus_ATCC_1997 TLLLAPFGYLPVDASRYWFITINTIAIVIAAYFLLRLFNFTLASVAAPAL
MSMEG_2785|M.smegmatis_MC2_155 TLLLAPFGYLPVDASRYWYISFNVLAFLIAAYLMLRIFDYTLSSVAAPAL
MMAR_2043|M.marinum_M TLLMAPFGYLPFAPSRYLFISINTAAILLAAYLLLRLFNFTLTSVAAPAL
MUL_3307|M.ulcerans_Agy99 TLLMAPFGYLPFAPSRYLFISINTAAILLAAYLLLRLFNFTLTSVAAPAL
Mb2692|M.bovis_AF2122/97 TLLMAPFGYLPFAPSRYLFISINTAAILVAAYLLLRMFNFTLTSVAAPAL
Rv2673|M.tuberculosis_H37Rv TLLMAPFGYLPFAPSRYLFISINTAAILVAAYLLLRMFNFTLTSVAAPAL
MLBr_01338|M.leprae_Br4923 TLLMAPFGYLPFAPSRYLFILINTGAILIAWYLILRLFKYTLSSVAAPTL
MAV_3565|M.avium_104 TLLMAPFGYLPEYPSRYLFILINTVAILIAWYLLLRLFGYTLSSVAAPAL
**::******. ** :* :*. *:::* *::**:* :**:*****:*
Mflv_3915|M.gilvum_PYR-GCK LLAMFCTESVTNTLVFGNINGVLLLLEVLFFRWLLDG----NRNHEWLAG
Mvan_2488|M.vanbaalenii_PYR-1 LLAMFCTESVTNTLVFGNINGVILLLEVLFFRWLLDG----NRSHEWWAG
TH_0624|M.thermoresistible__bu LLAMFCTETVTNTLVFTNINGILLLLEVLFLRWLLDG----EKRHEWLAG
MAB_2978|M.abscessus_ATCC_1997 LLAMFCTESVTNTLVFTNINGVVLLLEVLFFRWLLHG----TKKSEWLAG
MSMEG_2785|M.smegmatis_MC2_155 VLAMFCTESVTNTLVFTNINGCMLLGAVLFFRWLLKG----GRNAELLAG
MMAR_2043|M.marinum_M VLAMFATETVTNTLVFTNINGCILLLEVLFLRWLLDG----RVSHQWWAG
MUL_3307|M.ulcerans_Agy99 VLAMFTTETVTNTLVFTNINGCILLLEVLFLRWLLDG----RVSHQWWAG
Mb2692|M.bovis_AF2122/97 ILAMFATETVTNTLVFTNINGCILLLEVLFLRWLLDG----RASRQWCGG
Rv2673|M.tuberculosis_H37Rv ILAMFATETVTNTLVFTNINGCILLLEVLFLRWLLDG----RASRQWCGG
MLBr_01338|M.leprae_Br4923 LLAMFCTETVTSTLVFTNINGCIMLLEVLFLWWLINGSEPKTVSQQWWAG
MAV_3565|M.avium_104 LLAMFCTETVTNTLVFTNINGCVLLAEMLFLRWLLDG----RVGRQWWAG
:**** **:**.**** **** ::* :**: **:.* : .*
Mflv_3915|M.gilvum_PYR-GCK ISIGLTLVVKPLLAPLLLLPLLNRQWRALVAAFVVPLVFNAAAWP-----
Mvan_2488|M.vanbaalenii_PYR-1 VSIGLTLVVKPLLAPLLLLPLLNRQWRALVTAFAVPVAFNLAAWP-----
TH_0624|M.thermoresistible__bu VSIGLTLVVKPLLAPLLLLPVLNRQWRALVTAFAVPLAFNAAAWP-----
MAB_2978|M.abscessus_ATCC_1997 VAIGLTLVIKPLLAPLLLLPLLNRQWRVFAAAFGIPAFFNLVALYGPRKV
MSMEG_2785|M.smegmatis_MC2_155 AAIGLTLVVKPSLAPLLLLPVLNRQFYTLITAFGVPLVFNIAAWP-----
MMAR_2043|M.marinum_M IAIGFTLVLKPLLGPLLLLPLLNRQWRTLVTAIVIPIVINLAALP-----
MUL_3307|M.ulcerans_Agy99 IAIGFTLVLKPLLGPLLLLPLLNRQWRTLVTAIVIPIVINLAALP-----
Mb2692|M.bovis_AF2122/97 LAIGLTLVLKPLLGPLLLLPLLNRQWRALVAAVVVPVVVNVAALP-----
Rv2673|M.tuberculosis_H37Rv LAIGLTLVLKPLLGPLLLLPLLNRQWRALVAAVVVPVVVNVAALP-----
MLBr_01338|M.leprae_Br4923 GAIGLTLVLKPLLGPLLCLPLLNRQWQALVPAIALPVVINLAALP-----
MAV_3565|M.avium_104 VAIGLTLTLKPVLGPLLLLPLLNRQWRALVPAFAIPILINAAAWP-----
:**:**.:** *.*** **:****: .: .*. :* .* .*
Mflv_3915|M.gilvum_PYR-GCK LISDPMNFVTRTVPYILSTRDYFNSSILGNGLYYGLPMWLIMTLRVTIIV
Mvan_2488|M.vanbaalenii_PYR-1 LISDPMNFVTRTVPYIFSTRDYFNSSILGNGLYYGLPMWLIMSLRIAFVV
TH_0624|M.thermoresistible__bu LVADPMNFVTRTVPYILSTRDYFNSSILGNGVYYGLPTWLIILLRIGFGL
MAB_2978|M.abscessus_ATCC_1997 RIVDGWDYLRRTVKYLGETRDYFNSSIAGNGLYYGLPEPLIAFLRIAFLA
MSMEG_2785|M.smegmatis_MC2_155 LVPDPMNFVRHTVPYIMSTRDYFNSSIVGNGIYYGLPMWLILLLRVVFLL
MMAR_2043|M.marinum_M LINHPMDFFTRTLPYILGTRDYFNSSIQGNGIYFGLPTWLILFLRLLFTA
MUL_3307|M.ulcerans_Agy99 LINHPMDFFTRTLPYILGTRDYFNSSIQGNGIYFGLPTWLILFLRLLFTA
Mb2692|M.bovis_AF2122/97 LVSDPMSFFTRTLPYILGTRDYFNSSILGNGVYFGLPTWLILFLRILFTA
Rv2673|M.tuberculosis_H37Rv LVSDPMSFFTRTLPYILGTRDYFNSSILGNGVYFGLPTWLILFLRILFTA
MLBr_01338|M.leprae_Br4923 LVSHPMDFFTRTVPYILGTRDYFNSSIEGNGVYFGLPTWLIVFLRLLFTV
MAV_3565|M.avium_104 LVSDPMDFVTRTLPYIGGVRDYFNSSIEGNGVYFGLPTWLIVFMRILFTL
: . .:. :*: *: .******** ***:*:*** ** :*: :
Mflv_3915|M.gilvum_PYR-GCK LGAISLWLLYRHYRTRDPFFWMLTSSGVLLTTSWLALSLGQGYYSIALFP
Mvan_2488|M.vanbaalenii_PYR-1 LGAAALWLLYRYYRTRDPFFWMLTSSGVVLVTSWLVLSLGQGYYSIVLFP
TH_0624|M.thermoresistible__bu LAVVALVLLYRYYRTRDPLLWMTTSAGVLLIASWLVLSLGQGYYSMMLFP
MAB_2978|M.abscessus_ATCC_1997 IALVSVWLLYKYYRRRDPRFWALTTGGVLLTASFLLLGLGQGYYSMALFP
MSMEG_2785|M.smegmatis_MC2_155 LAVGSLWLLYRYYRERDPRFWLLTSSGVLLTASFLLLSLGQGYYSTMLFP
MMAR_2043|M.marinum_M IAIASLWLLYRYYRTRDPLFWFTTSSGVLLLWSWLAMSLAQGYYSMMLFP
MUL_3307|M.ulcerans_Agy99 IAIASLWLLYRYYRTRDPLFWFTTSSGVLLLWSWLAMSLAQGYYSMMLFP
Mb2692|M.bovis_AF2122/97 ITFGALWLLYRYYRTGDPLFWFTTSSGVLLLWSWLVMSLAQGYYSMMLFP
Rv2673|M.tuberculosis_H37Rv ITFGALWLLYRYYRTGDPLFWFTTSSGVLLLWSWLVMSLAQGYYSMMLFP
MLBr_01338|M.leprae_Br4923 LAICSLWLLNRYYRTRDPLFWFTCSTGVLLLWSWLVLPLAQGYYSMMLFP
MAV_3565|M.avium_104 IAIGALWLLYRYYRTRDPRFWFTNSAGVLLLWSWLVLPLAQGYYSMMLFP
: :: ** ::** ** :* : **:* *:* : *.***** ***
Mflv_3915|M.gilvum_PYR-GCK FLMTVVLPNSVIRSWVAWLAAYGFLTMDRWLLWQ---LPSTGRAFEYLKI
Mvan_2488|M.vanbaalenii_PYR-1 FLMTVVLPNSVIRNWVAWLAAYGFLTMDRWLLWQ---LPTTGRALEYLKI
TH_0624|M.thermoresistible__bu FLMTVVLPNSVIRNWVAWLAVYGFMTMDRWLIVRPVEMLTTGRFLEYMKI
MAB_2978|M.abscessus_ATCC_1997 FVMTIVLPNSVLRNWPAWLAVYGFFSMDRWLLGH---WPTTGRALEYLKI
MSMEG_2785|M.smegmatis_MC2_155 FLMTVVLPNSVLRNWPAWLAIYGFMTMDRWLLGH---WPTTGRFLEYMKI
MMAR_2043|M.marinum_M FLMTITLRNSVIRNWPAWLGIYGFMTLDRWLLFN---WMRWGRALEYLKI
MUL_3307|M.ulcerans_Agy99 FLMTITLRNSVIRNWPAWMGIYGFMTLDRWLLFN---WMRWGRALEYLKI
Mb2692|M.bovis_AF2122/97 FLMTVVLPNSVIRNWPAWLGVYGFMTLDRWLLFN---WMRWGRALEYLKI
Rv2673|M.tuberculosis_H37Rv FLMTVVLPNSVIRNWPAWLGVYGFMTLDRWLLFN---WMRWGRALEYLKI
MLBr_01338|M.leprae_Br4923 FLMTVVLPNSLIRNWPAWLGIYGFLTLDRWLLFN---WMRYGRALEYLKI
MAV_3565|M.avium_104 FLMTVVLPNSLIRNWPAWLGIYGFMTLDRWLLFN---WMRYGRALEYLKI
*:**:.* **::*.* **:. ***:::****: . ** :**:**
Mflv_3915|M.gilvum_PYR-GCK TYGWSLMMVVVFAVLLFRYLDARQDNRLDEGIDPAWMK----QLTPAAKP
Mvan_2488|M.vanbaalenii_PYR-1 TFGWSLMSVVVFSVLLFRYLDAKDDGRLDEGIDPPWMK----QLTPADKA
TH_0624|M.thermoresistible__bu TYGWSLMLIVVTTVLYFRYLDAKADGRLDEGIDPPWLQKSKAEPVPATTS
MAB_2978|M.abscessus_ATCC_1997 TYGWCLLLIVVMMVLVLRYRDARAAGTLDQGLDPEWMRAQTGTKESATDD
MSMEG_2785|M.smegmatis_MC2_155 TYGWSLMLVVVFCVLYFRYLDAKQDGRLDQGIDPPWMAR---ERTPAPV-
MMAR_2043|M.marinum_M TYGWSLLLIVTFTVLYFRYLDAKAEDRLDSGIDPAWLTA---ERERASVD
MUL_3307|M.ulcerans_Agy99 TYGWSLLLIVTFTVLYFRYLDAKAEDRLDSGIDPAWLTA---ERERASVD
Mb2692|M.bovis_AF2122/97 TYGWSLLLIVTFTVLYFRYLDAKADNRLDGGIDPAWLTP---EREGQR--
Rv2673|M.tuberculosis_H37Rv TYGWSLLLIVTFTVLYFRYLDAKADNRLDGGIDPAWLTP---EREGQR--
MLBr_01338|M.leprae_Br4923 TYGWSLLLIVVSTVLCFRYLDAKAENRLDHGIDPAWLTA---ERERASVN
MAV_3565|M.avium_104 TYGWSLLLIVVFSVLCYRYLDAKAEDRLDRGIDPAWLTA---GRE-----
*:**.*: :*. ** ** **: . ** *:** *:
Mflv_3915|M.gilvum_PYR-GCK SE------------------
Mvan_2488|M.vanbaalenii_PYR-1 AE------------------
TH_0624|M.thermoresistible__bu ASGS----------------
MAB_2978|M.abscessus_ATCC_1997 DIERPEGESDRRAVAGATDS
MSMEG_2785|M.smegmatis_MC2_155 --------------------
MMAR_2043|M.marinum_M A-------------------
MUL_3307|M.ulcerans_Agy99 A-------------------
Mb2692|M.bovis_AF2122/97 --------------------
Rv2673|M.tuberculosis_H37Rv --------------------
MLBr_01338|M.leprae_Br4923 A-------------------
MAV_3565|M.avium_104 --------------------