For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RQDSGIGVIDKAVGLLRATAESPCGLAELCERTGLPRATAYRLAAALEVHRLLGRDEDGRWGLGPAITEL AAHVDDPLLTAGATVLPRLRETTGESVQLYRREGTSRVCVAALEPPAGLRDTVPVGARLPMTAGSGAKVL LAHSDPATQQAVLPTAKFTDRVLAEVRRRGWAQSVAEREPGVASVSAPVRDGRGVVVAAISVSGPIDRIG RRPGARWAADLLAAADELARRL*
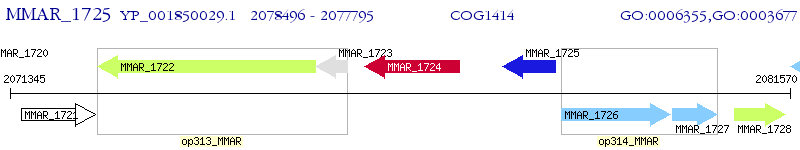
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1725 | - | - | 100% (233) | transcriptional regulatory protein |
| M. marinum M | MMAR_4423 | - | 4e-17 | 30.58% (242) | transcriptional regulatory protein |
| M. marinum M | MMAR_4779 | - | 6e-15 | 31.41% (191) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3013 | - | 1e-109 | 84.98% (233) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_4228 | - | 1e-107 | 82.40% (233) | regulatory protein, IclR |
| M. tuberculosis H37Rv | Rv2989 | - | 1e-109 | 84.98% (233) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3295 | - | 1e-102 | 78.97% (233) | IclR family transcriptional regulator |
| M. avium 104 | MAV_3839 | - | 1e-104 | 85.45% (220) | transcriptional regulator, IclR family protein |
| M. smegmatis MC2 155 | MSMEG_2386 | - | 1e-110 | 84.12% (233) | transcriptional regulator, IclR family protein |
| M. thermoresistible (build 8) | TH_2042 | - | 1e-110 | 84.12% (233) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. ulcerans Agy99 | MUL_1967 | - | 1e-128 | 99.14% (233) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_2134 | - | 1e-110 | 83.69% (233) | regulatory proteins, IclR |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1725|M.marinum_M MRQDSGIGVIDKAVGLLRATAESPCGLAELCERTGLPRATAYRLAAALEV
MUL_1967|M.ulcerans_Agy99 MRQDSGIGVIDKAVGLLRATAKSPCGLAELCERTGLPRATAYRLAAALEV
Mb3013|M.bovis_AF2122/97 MRQHSGIGVLDKAVGVLHAVAESPCGLAELCDRTDLPRATAYRLAAALEV
Rv2989|M.tuberculosis_H37Rv MRQHSGIGVLDKAVGVLHAVAESPCGLAELCDRTDLPRATAYRLAAALEV
MAV_3839|M.avium_104 -------------MGVLHTVAQSPCGLAELCERTELPRATAYRLAAALEV
MSMEG_2386|M.smegmatis_MC2_155 MRNDSGIGVLDKAVGVLHTVAESPCGLAELCERTGLPRATAHRLAAGLET
TH_2042|M.thermoresistible__bu MRKDSGIGVLDKAVSVLHSVAESPCGLAELCERTGLPRATAHRLAAGLEV
Mflv_4228|M.gilvum_PYR-GCK MRQNSGIGVLDKAITVLHAVAESPCGLADLCERTDLPRATAHRLAVGLEV
Mvan_2134|M.vanbaalenii_PYR-1 MRQNSGIGVLDKAVTVLHAVAESPCGLADLCERTNLPRATAHRLAVGLEV
MAB_3295|M.abscessus_ATCC_1997 MRQHSGIGVLDKAVQVLTAIGDSPCGLAELCERTGLPRATAHRLAAGLEV
: :* : ..******:**:** ******:***..**.
MMAR_1725|M.marinum_M HRLLGRDEDGRWGLGPAITELAAHVDDPLLTAGATVLPRLRETTGESVQL
MUL_1967|M.ulcerans_Agy99 HRLLGRDEDGRWGLGPAITELAAHVDDPLLTAGATVLPRLCETTGESVQL
Mb3013|M.bovis_AF2122/97 HRLLGRGQDGHWRLGPAITELATHVDDPLLVACAAVLPQLRDATGESVQV
Rv2989|M.tuberculosis_H37Rv HRLLGRGQDGHWRLGPAITELATHVDDPLLVACAAVLPQLRDATGESVQV
MAV_3839|M.avium_104 HRLLARGDDGRWRLGPAVTELAAHVNDPLLAAGSAVLPLLRETTGESVQL
MSMEG_2386|M.smegmatis_MC2_155 HRLLARDADGRWRLGPALTELAAHVNDPLLSAGAAVLPRLREITGESVQL
TH_2042|M.thermoresistible__bu HRLLARDGDGRWCLGPAVTELAARVDDPLLAAAAVVLPRLREITGESVQL
Mflv_4228|M.gilvum_PYR-GCK HRLLARDGDGLWRLGPALTELAGHVNDSLLAAGAVVLPRLRELTGESVQL
Mvan_2134|M.vanbaalenii_PYR-1 HRLLAREGDGQWRLGPALTELAGHVNDPLLAAGAVVLPRLRELTGESVQL
MAB_3295|M.abscessus_ATCC_1997 HRLLSRDGDGQWQLGAGLTELAAHVNDPLLSASAQVLPKLREITGESVQL
****.* ** * **..:**** :*:*.** * : *** * : ******:
MMAR_1725|M.marinum_M YRREGTSRVCVAALEPPAGLRDTVPVGARLPMTAGSGAKVLLAHSDPATQ
MUL_1967|M.ulcerans_Agy99 YRREGTSRVCVAALEPPAGLRDTVPVGARLPMTAGSGAKVLLAHSDPATQ
Mb3013|M.bovis_AF2122/97 YRREGTSRVCVAALEPAAGLRDTVPVGARLPMTAGSGAKVLLAHTDAATQ
Rv2989|M.tuberculosis_H37Rv YRREGTSRVCVAALEPAAGLRDTVPVGARLPMTAGSGAKVLLAHTDAATQ
MAV_3839|M.avium_104 YRREGTDRVCVAALEPAAGLRDTVPVGARLPMTAGSGAKVLLAHSDAATQ
MSMEG_2386|M.smegmatis_MC2_155 YRREGLSRVCVVALEPPAGLRDTVPVGTQLPMTAGSGAKVLLAYADAATQ
TH_2042|M.thermoresistible__bu YRREGTSRICVAALEPPAGLRDTVPVGTRLPMTAGSGAKVLLAYADSTTQ
Mflv_4228|M.gilvum_PYR-GCK YRREGTSRICVAALEPPAGLRDTVPVGTRLSMTAGSGAKVLLAYADTHTQ
Mvan_2134|M.vanbaalenii_PYR-1 YRREGTTRICIAALEPPAGLRDTVPVGIRLPMTAGSGAKVLLAYADAATQ
MAB_3295|M.abscessus_ATCC_1997 YRREGTVRICVAALEPPAGLRDTVPVGSRLPMSAGSGAKVLLAHSDPGTQ
***** *:*:.****.********** :*.*:**********::*. **
MMAR_1725|M.marinum_M QAVLPTAKFTDRVLAEVRRRGWAQSVAEREPGVASVSAPVRDGRGVVVAA
MUL_1967|M.ulcerans_Agy99 QAVLPTAKFTDRVLAEVRRRGWAQSVAEREPGVASVSAPVRDGRGVVVAA
Mb3013|M.bovis_AF2122/97 AAVLPKAVFSARALAEVCRRGWAQSVAEREPGVASVSAPVRDGRGVVIAA
Rv2989|M.tuberculosis_H37Rv AAVLPKAVFSARALAEVCRRGWAQSVAEREPGVASVSAPVRDGRGVVIAA
MAV_3839|M.avium_104 KAVLANAKFTERTLAEVRRRGWAQSVAEREPGVASVSAPVRDSRGAVIAA
MSMEG_2386|M.smegmatis_MC2_155 QAVLPSAKFTERTLAEVRKRGWAQSAAEREPGVASVSAPVRDSRGQVIAA
TH_2042|M.thermoresistible__bu QAVLSNAKFSERALAEVHKRGWAQSVAEREPGVASVSAPVRDGRGTVVAA
Mflv_4228|M.gilvum_PYR-GCK QAVLPNAKFTDRTLAEVRKRGWAQSAAEREPGVASVSAPVRDGRGAVIAA
Mvan_2134|M.vanbaalenii_PYR-1 QAVLPGAKFTDRTLAEVRRRGWAQSAAEREPGVASVSAPVRDGRGAVVAA
MAB_3295|M.abscessus_ATCC_1997 AAVLPGATFTERALADVRKRGWAQSAAEREAGVASVSAPVRDRHGNVIAA
***. * *: *.**:* :******.****.*********** :* *:**
MMAR_1725|M.marinum_M ISVSGPIDRIGRRPGARWAADLLAAADELARRL
MUL_1967|M.ulcerans_Agy99 ISVSGPIDRIGRRPGARWAADLLAAADELARRL
Mb3013|M.bovis_AF2122/97 ISVSGPIDRMGRRPGVRWAADLLSAADALTRRL
Rv2989|M.tuberculosis_H37Rv ISVSGPIDRMGRRPGVRWAADLLSAADALTRRL
MAV_3839|M.avium_104 ISVSGPIDRIGRRPGARWAADLVSAAEALTRRL
MSMEG_2386|M.smegmatis_MC2_155 VSVSGPIDRMGRRPGARWAADLLAAAEALTRRL
TH_2042|M.thermoresistible__bu VSVSGPIDRIGRRPGARWAADLLAAAEALTRRL
Mflv_4228|M.gilvum_PYR-GCK VSVSGPIDRMGRRPGARWAADLVAAADALTRRL
Mvan_2134|M.vanbaalenii_PYR-1 VSVSGPIDRMGRRPGARWAADLVAAADALTRRL
MAB_3295|M.abscessus_ATCC_1997 ISVSGPIDRMGRRPGARWAADLVAASEAITARL
:********:*****.******::*:: :: **