For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGIALTDDHRELAEVARAFLTSQKARWAARSLLDSPDESRPPFWQDMVELGWLGLHIDERHGGSGYGLPE LVVVVEELGRAVAPGPFVPTVIASAVLAKEGSDEQQTRLLPGLIDGTVTAGIGLDGNLRVENGVASGDAG IVLGAGLAELLLVAAGDEVLVLDRGRSGVSVEVPDNFDPTRRSGRVRLHEVKVSAQDLLPGARAAALARA RTVLAAEAVGGAADCVDAAVAYAKVRQQFGRTIATFQAVKHHCANMLVAAESGIAAVWDAARAATEDEEQ FQLIAAVAALLAFPAYARNAELNIQVHGGIGFTWEHDAHLHLRRALVDAALLGAEAPARDVFERTAAGAT RENSLDLPAEAEELRTQIRADAVEIAALDKAAQLDKLIETGYVMPHWPKPWGRAADAVEQLVIEEEFRKA AIKRPDYSITGWVILTLIQHGTAWQIERFVEKALRQEEIWCQLFSEPEAGSDAASVKTRATRVDGGWKIN GQKVWTSGAQYCARGLATVRTDPDVPKHAGITTVIIDMKSPGVEVRPLRQITGGSEFNEVFFNDVFVPDE DVVGTPNSGWTVARATLGNERVSIGGSGGYYEGLAAKLVKLAQNGADRLAGAEIRVGYFLAEDHALRLLN LRRAARSVEGAGPGPEGNVTKLKLAEHMVEGAAISAALLGPEIALLDGPGAVIGRMVMGARGMAIAGGTS EVTRNQIAERILGMPRDPLIN
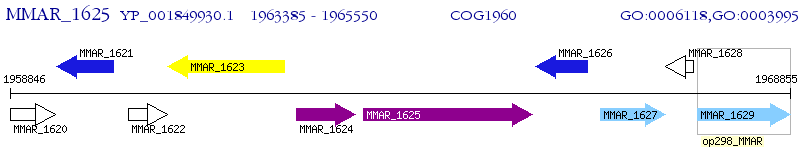
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1625 | fadE22 | - | 100% (721) | acyl-CoA dehydrogenase FadE22 |
| M. marinum M | MMAR_0530 | fadE6 | e-152 | 41.85% (736) | acyl-CoA dehydrogenase FadE6 |
| M. marinum M | MMAR_5068 | fadE34 | e-137 | 42.11% (722) | acyl-CoA dehydrogenase FadE34 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3087c | fadE22b | 0.0 | 85.83% (600) | acyl-CoA dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0441 | - | 0.0 | 53.79% (725) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3061c | fadE22 | 0.0 | 85.02% (721) | acyl-CoA dehydrogenase FADE22 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 7e-19 | 26.39% (379) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0580 | - | 1e-139 | 41.87% (695) | acyl-CoA dehydrogenase FadE |
| M. avium 104 | MAV_3935 | - | 0.0 | 85.85% (721) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0603 | - | 1e-149 | 41.86% (743) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_3278 | - | 0.0 | 77.21% (724) | PUTATIVE acyl-CoA dehydrogenase FadE22 |
| M. ulcerans Agy99 | MUL_3411 | fadE22 | 0.0 | 99.03% (721) | acyl-CoA dehydrogenase FadE22 |
| M. vanbaalenii PYR-1 | Mvan_0226 | - | 0.0 | 52.28% (725) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0441|M.gilvum_PYR-GCK MSKTAS----ASKLAITDDHQDLAAAVTGQLDRLKALVLARATLDAADTD
Mvan_0226|M.vanbaalenii_PYR-1 MSKTVSATVTASRLAITDDHQDLAAAVSGQLTRLQTPAKARAALAG----
Mb3087c|M.bovis_AF2122/97 --------------------------------------------------
Rv3061c|M.tuberculosis_H37Rv -----------MGIALTDDHRELSGVARAFLTSQKVRWAARASLDA----
MMAR_1625|M.marinum_M -----------MGIALTDDHRELAEVARAFLTSQKARWAARSLLDS----
MUL_3411|M.ulcerans_Agy99 -----------MGIALTDDHRELAEVARAFLTSQKARWAARSLLDS----
MAV_3935|M.avium_104 -----------MGIALTDDHRELAEVARGFLTSQKARWAARSLLDA----
TH_3278|M.thermoresistible__bu -----------MAIALTDDHRELAEVARSFLTSQQARAAARALLDA----
MSMEG_0603|M.smegmatis_MC2_155 -----------MPIAILSEHIDLADSVKSFVARVAPSDVLHEALETPLET
MAB_0580|M.abscessus_ATCC_1997 -----MSVPSGLRASGSSEVQEAARDAVRQWARSAAVIESVRKMES----
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_0441|M.gilvum_PYR-GCK GASNPSDLWSAATGLGWTGLALAEDVGGSGFGLAELAVVVECLGHQLAPG
Mvan_0226|M.vanbaalenii_PYR-1 GSTHPADIWSAATELGWTGLALPEDVGGSGFGLEELAVVIECLGSQLAPG
Mb3087c|M.bovis_AF2122/97 --------------------------------------------------
Rv3061c|M.tuberculosis_H37Rv AGDARPPFWQNLAELGWLGLHIDERHGGSGYGLSELVVVIEELGRAVAPG
MMAR_1625|M.marinum_M PDESRPPFWQDMVELGWLGLHIDERHGGSGYGLPELVVVVEELGRAVAPG
MUL_3411|M.ulcerans_Agy99 PDESRPPFWQDMVELGWLGLHIDERHGGSGYGLPELVVVVEELGRAVAPG
MAV_3935|M.avium_104 TDEPRPGFWPNLVELGWLGLHIDEEYGGSGFGLPELVVVVEELGRAVAPG
TH_3278|M.thermoresistible__bu DDEARPPFWSELAGLGWLGLHISEKHGGSGYGLPELVVVVEELGRAVAPG
MSMEG_0603|M.smegmatis_MC2_155 PRANPPSYWKAAADQGLQGVHLSESVGGQGFGILELAIVLAEFGYGAVPG
MAB_0580|M.abscessus_ATCC_1997 EPDGWGPAYVGLAELGIFGVAVPEAAGGSGAGFDDMIAMVDEAAASLVPG
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_0441|M.gilvum_PYR-GCK PFLTSVSAAVVIDRCAPDSVRAELLPGSASGDTVAALGISGTVAVGAD--
Mvan_0226|M.vanbaalenii_PYR-1 PFLSSAAASVVIDRSAPDSVRAELLPVLAGGETVAALGVSGSVVVGTD--
Mb3087c|M.bovis_AF2122/97 ------------------------------------MGLDSQVQVTD---
Rv3061c|M.tuberculosis_H37Rv LFVPTVIASAVVAKEGTDDQRARLLPALIDGTLTAGVGLDSQVQVTD---
MMAR_1625|M.marinum_M PFVPTVIASAVLAKEGSDEQQTRLLPGLIDGTVTAGIGLDGNLRVEN---
MUL_3411|M.ulcerans_Agy99 PFVPTVIASAVLAKEGSDEQQTRLLPGLIDGTVTAGIGLDGNVRVEN---
MAV_3935|M.avium_104 PFVPTVIASAVIAKNGTAEQKSRLLPGLIDGTVTAGIGLDSRVRLGD---
TH_3278|M.thermoresistible__bu PFVPTVVASAVLADAGSDEQRDRLLPGLVDGSVVAAVGLGGDLRVEGPVE
MSMEG_0603|M.smegmatis_MC2_155 PFVPSAIASALISAHDPD---AKVLSELASGDAIAAYALDSTLTATRQGD
MAB_0580|M.abscessus_ATCC_1997 PVATSALVAVVLAADGHE----ELVPPLAAGERTAGLALTGNLAIADG--
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_0441|M.gilvum_PYR-GCK -NTATGECATVLGAPDASLLVL----VAGDDVVIVDAG----DDGVTVTR
Mvan_0226|M.vanbaalenii_PYR-1 -LIVSGESPVVLAAPDASVLVL----TAGEDVVILDAGPASAAGGVTVTP
Mb3087c|M.bovis_AF2122/97 -GVADGEAGIVLGAGLAELLLV----AAGDDVLVLERG----RKGVSVDV
Rv3061c|M.tuberculosis_H37Rv -GVADGEAGIVLGAGLAELLLV----AAGDDVLVLERG----RKGVSVDV
MMAR_1625|M.marinum_M -GVASGDAGIVLGAGLAELLLV----AAGDEVLVLDRG----RSGVSVEV
MUL_3411|M.ulcerans_Agy99 -GVASGDAGIVLGAGLAELLLV----AAGDDVLVLDRG----RSGVSVEV
MAV_3935|M.avium_104 -GVADGEAGILLGAGLAGLLLI----AAGDDVLVLERD----RDGVSVDV
TH_3278|M.thermoresistible__bu AATASGTAGVVLGAGRADLLLL----RCGDDVLIVDRT----APGVTVEV
MSMEG_0603|M.smegmatis_MC2_155 GLVIRGEVRAVPAAAQASILVLPVAIDSGEEWVVLDAD------QLEIEP
MAB_0580|M.abscessus_ATCC_1997 --LASGVLPAVLGGTADGVLLA----PVPDGWVLVDCA----EAGVAVEA
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_0441|M.gilvum_PYR-GCK LDCLDTTRSIGSVTLDGVAVPADRILRG-AARDARTVFRTLTAAEAVGVS
Mvan_0226|M.vanbaalenii_PYR-1 LDCLDPTRSIGSVTLRDVPVAADRVLRG-AARNARTVFRILAAAEAVGVS
Mb3087c|M.bovis_AF2122/97 PENFDPTRRSGRVRLDNVRVTTDDILLG-AYESALARARTLLAAEAVGGA
Rv3061c|M.tuberculosis_H37Rv PENFDPTRRSGRVRLDNVRVTTDDILLG-AYESALARARTLLAAEAVGGA
MMAR_1625|M.marinum_M PDNFDPTRRSGRVRLHEVKVSAQDLLPG-ARAAALARARTVLAAEAVGGA
MUL_3411|M.ulcerans_Agy99 PDNFDPTRRSGRVRLHEVKVSAQDLLSG-ARAAALARARTVLAAEAVGGA
MAV_3935|M.avium_104 PDNFDPTRRSGRVRLDNVRVSDADVLAG-ARPSALARARTLLAAEAVGGA
TH_3278|M.thermoresistible__bu PTNLDPTRRSGRVTLTGVAVT--DVLPG-AAETALALARTLFAAEAAGGA
MSMEG_0603|M.smegmatis_MC2_155 VRSVDPLRPLAHVRANAVEVADERVLSNLSRPLARALMSTLLSAESIGVA
MAB_0580|M.abscessus_ATCC_1997 KKATDFSRPWVTVTLHGAAVRPVGLPAA----VIADLSVVTLSAEAVGVA
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_0441|M.gilvum_PYR-GCK WAALDMAVEYAKVREQFGRTIGTFQAVKHHAANMLVSAETATAAVWDAAR
Mvan_0226|M.vanbaalenii_PYR-1 WAALDMAVGYAKVREQFGRTIGTFQAVKHHAANMLVAAEVATAATWDAAR
Mb3087c|M.bovis_AF2122/97 ADCVDSAVAYAKVRQQFGRTIATFQAVKHHCANMLVAAESAIAAVWDAAR
Rv3061c|M.tuberculosis_H37Rv ADCVDSAVAYAKVRQQFGRTIATFQAVKHHCANMLVAAESAIAAVWDAAR
MMAR_1625|M.marinum_M ADCVDAAVAYAKVRQQFGRTIATFQAVKHHCANMLVAAESGIAAVWDAAR
MUL_3411|M.ulcerans_Agy99 ADCVDAAVAYAKVRQQFGRTIATFQAVKHHCANMLVAAESGIAAVWDAAR
MAV_3935|M.avium_104 SDCVDAAVAYAKVRQQFGRTIATFQAVKHHCANMLVGAESGIAAVWDAAR
TH_3278|M.thermoresistible__bu VECVETAVEYAKVRRQFGRVIGTFQAVKHHCANMAVAAESAIATVWDAAR
MSMEG_0603|M.smegmatis_MC2_155 RWATDTAAGYAKIREQFGRPIGQFQAVKHKCANMVAVTERATAAVWDAAR
MAB_0580|M.abscessus_ATCC_1997 RWALQTAVEYAKVREQFGKQIGSFQAIKHMCAEMLCRVEQADVAVWDAAG
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_0441|M.gilvum_PYR-GCK ADDV------DSAWF--AAAVAAALAIKAQVFNAQNNIQLHGGIGFTWEH
Mvan_0226|M.vanbaalenii_PYR-1 ADDL------DSAWF--AGAVAAALAIRAQVFNAQNNIQLHGGIGFTWEH
Mb3087c|M.bovis_AF2122/97 AAAE------DEEQFRLAAAVAAALAFPAYARNAELNIQVHGGIGFTWEH
Rv3061c|M.tuberculosis_H37Rv AAAE------DEEQFRLAAAVAAALAFPAYARNAELNIQVHGGIGFTWEH
MMAR_1625|M.marinum_M AATE------DEEQFQLIAAVAALLAFPAYARNAELNIQVHGGIGFTWEH
MUL_3411|M.ulcerans_Agy99 AATE------DEEQFQLIAAVAALLAFPAYARNAELNIQVHGGIGFTWEH
MAV_3935|M.avium_104 AAGE------DEDQFHLIAAVAAALAFPAYVRNAELNIQVHGGIGFTWEH
TH_3278|M.thermoresistible__bu AAAG------DPAEFRLMAAAAAALAFPAYVHNAELNIQVHGGIGYTWEH
MSMEG_0603|M.smegmatis_MC2_155 ALDEVREKGTEGTHFGFAAAVAATLAPAAAQQCTQDCIQVHGGIGFTWEH
MAB_0580|M.abscessus_ATCC_1997 CAQDVLANKGDSQQLSLAAAVAAAVAVDAAVANTKDCIQILGGIGFTWEH
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_0441|M.gilvum_PYR-GCK DAHLYLRRARTLAALMADGVDPLIDIVDGQRTGRAHGASFTLPPEADEYR
Mvan_0226|M.vanbaalenii_PYR-1 DSHLYLRRARTLAALMADGVDPLLDIVDGQRNGRAHGASFTLPPESEEYR
Mb3087c|M.bovis_AF2122/97 DAHLHLRRALVTVGLFG-GDAPVRDVFERTAAGVTRAISLDLPAQAEELR
Rv3061c|M.tuberculosis_H37Rv DAHLHLRRALVTVGLFG-GDAPVRDVFERTAAGVTRAISLDLPAQAEELR
MMAR_1625|M.marinum_M DAHLHLRRALVDAALLG-AEAPARDVFERTAAGATRENSLDLPAEAEELR
MUL_3411|M.ulcerans_Agy99 DAHLHLRRALVDAALLG-ADVPARDVFERTAAGATRENSLDLPAEAEELR
MAV_3935|M.avium_104 DAHLHLRRAVVTAALFG-GDAPAADVFERTAAGTVRDNSLDLPPEAEELR
TH_3278|M.thermoresistible__bu DAHLHLRRALAVRGVLG-GDGPAADVHALTAAGVTRENSLDLPPEAEEMR
MSMEG_0603|M.smegmatis_MC2_155 DTNVYYRRALLLAACFGRAADHPQQVVDTATATGMRPVDIDLDPETEKLR
MAB_0580|M.abscessus_ATCC_1997 DAHLYLRRAYALQSFVGKPAVWRRKAAALTIGGIRRSLTIDLG-EVESQR
MLBr_00737|M.leprae_Br4923 --------------MVGWSGNPLFDLFKLP--EEHNELRATIRALAEKEI
.. . . : :.
Mflv_0441|M.gilvum_PYR-GCK TQAREAAEAVRRLPAEQQRDYLVDSGYLVPHWPKPWG--RAADVLEQLVI
Mvan_0226|M.vanbaalenii_PYR-1 SQAREAAAKVRSLPAGQQRDFLVESGYLVPHWPQPWG--RAANVLEQLVI
Mb3087c|M.bovis_AF2122/97 ARIRSDAAEIAALEKDAQRDKLIETGYVMPHWPRPWG--RAAGAVEQLVI
Rv3061c|M.tuberculosis_H37Rv ARIRSDAAEIAALEKDAQRDKLIETGYVMPHWPRPWG--RAAGAVEQLVI
MMAR_1625|M.marinum_M TQIRADAVEIAALDKAAQLDKLIETGYVMPHWPKPWG--RAADAVEQLVI
MUL_3411|M.ulcerans_Agy99 TQIRADAVEIAALDKAAQLDKLIETGYVMPHWPKPWG--RAADAVEQLVI
MAV_3935|M.avium_104 TKIRADAAAIAALDKDAQLDKLIETGYVMPHWPKPWG--RAADAVEQLVI
TH_3278|M.thermoresistible__bu GRIRADIEEIAALDGNAQRDRLIETGYIMPHWPKPWG--RGADAVEQLVI
MSMEG_0603|M.smegmatis_MC2_155 DEIRAEVAALKAIPSEERTAAIAEGGWVQPHLPKPWG--RDATPVEQIII
MAB_0580|M.abscessus_ATCC_1997 AEVAGQVAEIASAPKAEHQVRLAEAGFLAPHWPAPYG--LGAGAAQQLLI
MLBr_00737|M.leprae_Br4923 APHAADVDQRARFP-EEALAALNASGFNAIHVPEEYGGQGADSVAACIVI
: *: * * :* ::*
Mflv_0441|M.gilvum_PYR-GCK EEEF--AAIERADMGITGWVALTIAQAGTDEQRERWVEPVLRGQVMWCQL
Mvan_0226|M.vanbaalenii_PYR-1 EEEF--AGVDRPDMGITGWVALTIAQAGTDDQRERWVEPVLRGQVMWCQL
Mb3087c|M.bovis_AF2122/97 EEEFSAAGIERPDYSITGWVILTLIQHGTPWQIERFVEKALRQQEIWCQL
Rv3061c|M.tuberculosis_H37Rv EEEFSAAGIERPDYSITGWVILTLIQHGTPWQIERFVEKALRQQEIWCQL
MMAR_1625|M.marinum_M EEEFRKAAIKRPDYSITGWVILTLIQHGTAWQIERFVEKALRQEEIWCQL
MUL_3411|M.ulcerans_Agy99 EEEFRKAAIKRPDYSITGWVILTLIQHGTAWQIERFVEKALRQEEIWCQL
MAV_3935|M.avium_104 EEEFRAAGIKRPDYGITGWVILTLIQHGTPWQIERFVEKALRKDEIWCQL
TH_3278|M.thermoresistible__bu DEEFARAGIERPDYSITGWVILTLIQHGTPSQIERFVEKALRQEEIWCQL
MSMEG_0603|M.smegmatis_MC2_155 AQEFTTGRVKRPQMGIASWIIPSIVAFGTDEQKQRFLPPTFRGEMIWCQL
MAB_0580|M.abscessus_ATCC_1997 DQELHAAGVNRPDLVIGWWAAPTILEHGTPEQIAQFVAPTLRGEIEWCQL
MLBr_00737|M.leprae_Br4923 EEVARVDASASLIPAVNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYA
: : : *: : : .
Mflv_0441|M.gilvum_PYR-GCK FSEPGAGSDAAAVRTSAKKVDGGWRVTGQKVWTSLAHMCQWGLATVRTDP
Mvan_0226|M.vanbaalenii_PYR-1 FSEPGAGSDAAAVRTSAKKVDGGWRVTGQKVWTSLAHLCQWGLATVRTDP
Mb3087c|M.bovis_AF2122/97 FSEPDAGSDAASVKTRATRVEGGWKINGQKVWTSGAQYCARGLATVRTDP
Rv3061c|M.tuberculosis_H37Rv FSEPDAGSDAASVKTRATRVEGGWKINGQKVWTSGAQYCARGLATVRTDP
MMAR_1625|M.marinum_M FSEPEAGSDAASVKTRATRVDGGWKINGQKVWTSGAQYCARGLATVRTDP
MUL_3411|M.ulcerans_Agy99 FSEPEAGSDAASVKTRATRVDGGWKINGQKVWTSGAQYCARGLATVRTDP
MAV_3935|M.avium_104 FSEPDAGSDAASIKTRATRVEGGWKINGQKVWTSGAHYCARGLATVRTDP
TH_3278|M.thermoresistible__bu FSEPDAGSDAAAVKTRAIRVDGGWKVTGQKVWTSGAHYCRRGLATVRTDP
MSMEG_0603|M.smegmatis_MC2_155 FSEPGAGSDLASLTTRAVKVDGGWRITGQKIWTTGAQFSAWGALLARTDP
MAB_0580|M.abscessus_ATCC_1997 FSEPGAGSDLAALRTKATRVDGGWKLDGQKVWNSKAQIADWAICLARTNP
MLBr_00737|M.leprae_Br4923 LSEREAGSDAASMRTRAKADGDDWILNGFKCWITNGGKSTWYTVMAVTDP
:** **** *:: * * ..* : * * * : . . . *:*
Mflv_0441|M.gilvum_PYR-GCK DAAKHAGVTMMAIDMNADGVTVNPLRGLTGD--SHFNEVFFDDVFVPDAD
Mvan_0226|M.vanbaalenii_PYR-1 DAPKHAGVTMMAIDMNAEGVTVNPLRGITGD--SHFNEVFFDDVFVPDAD
Mb3087c|M.bovis_AF2122/97 DAPKHAGITTVIIDMLAPGVEVRPLRQITGD--SEFNEVFFNDVFVPDED
Rv3061c|M.tuberculosis_H37Rv DAPKHAGITTVIIDMLAPGVEVRPLRQITGD--SEFNEVFFNDVFVPDED
MMAR_1625|M.marinum_M DVPKHAGITTVIIDMKSPGVEVRPLRQITGG--SEFNEVFFNDVFVPDED
MUL_3411|M.ulcerans_Agy99 DVPKHAGITTVIIDMKSPGVEVRPLRQITGG--SEFNEVFFNDVFVPDED
MAV_3935|M.avium_104 DAPKHAGITTVIVDMKAPEVEVRPLRQITGG--SDFNEVFFNDLFVPDED
TH_3278|M.thermoresistible__bu DAPKHAGITTMIIDMHAPGVEVRPLRMITGG--AEFNEVFFNDVFVPDED
MSMEG_0603|M.smegmatis_MC2_155 DAPKHQGITYFLLDMKSPGVEVKPLRELTGN--AMFNTVFIDDVFVPDDM
MAB_0580|M.abscessus_ATCC_1997 DVPKHKGITYFLLDMKTPGITVRPLREITGE--EMFNEVFLDGVIVPDEN
MLBr_00737|M.leprae_Br4923 DK-GANGISAFIVHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDR
* *:: . :. . : * . * . :::: :*.
Mflv_0441|M.gilvum_PYR-GCK VVGDVNKGWLVARATLGNERVSIGGGSGGVTGTTPDHLIKLLDSSPDRAP
Mvan_0226|M.vanbaalenii_PYR-1 VVGDVNKGWLVARATLGNERISIGGGSGGPVGTTPDHLIKLLDSSPATSA
Mb3087c|M.bovis_AF2122/97 VVGAPNSGWTVARATLGNERVSIGG-SGSYYEAMAAKLVQLVQRRSDAFA
Rv3061c|M.tuberculosis_H37Rv VVGAPNSGWTVARATLGNERVSIGG-SGSYYEAMAAKLVQLVQRRSDAFA
MMAR_1625|M.marinum_M VVGTPNSGWTVARATLGNERVSIGG-SGGYYEGLAAKLVKLAQNGADRLA
MUL_3411|M.ulcerans_Agy99 VVGTPNSGWTVARATLGNERVSIGG-SGGYYEGLAAKLVKLAQNGADRLA
MAV_3935|M.avium_104 VVGTPNSGWTVARATLGNERVSIGG-SGSFYEGLADQLVQLAQQRPDRLA
TH_3278|M.thermoresistible__bu VVGEPNTGWTVARATLGNERVSIGG-GSGARVFAATSLIPLVQRHADRLA
MSMEG_0603|M.smegmatis_MC2_155 VLGEVNRGWEVSRNTLTNERVSIGS-SEPPFLANLDQFVQFLRDG-QFDQ
MAB_0580|M.abscessus_ATCC_1997 VVGSVDDGWRLARTTLANERVAMAG--GGTLDEAMESLLALIGDT-ELDA
MLBr_00737|M.leprae_Br4923 IIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGE
::* . *: : ** : * ::.. :
Mflv_0441|M.gilvum_PYR-GCK MFVRQAGEVIAEIHSLRLLNLRRATRAISGAEPGPEGNVTKLVVAESGQH
Mvan_0226|M.vanbaalenii_PYR-1 MYVRQAGEVIAELHSLRLLNLRRATRAIAGAEPGPEGNVTKLLVAESGQR
Mb3087c|M.bovis_AF2122/97 GAPIRVGAFLAEDHALRLLNLRRAARSVEGAGPGPEGNITKLKVAEHMIE
Rv3061c|M.tuberculosis_H37Rv GAPIRVGAFLAEDHALRLLNLRRAARSVEGAGPGPEGNITKLKVAEHMIE
MMAR_1625|M.marinum_M GAEIRVGYFLAEDHALRLLNLRRAARSVEGAGPGPEGNVTKLKLAEHMVE
MUL_3411|M.ulcerans_Agy99 GAEIRVGYFLVEDHALRLLNLRRAARSVEGAGPGPEGNVTKLKLAEHMVE
MAV_3935|M.avium_104 GGKIRVGSYLAEETALRLLNLRRAARSVEGTGPGPEGNVTKLKLAEHMVE
TH_3278|M.thermoresistible__bu GAEVRVGTFLAEEHALRLLNLRRAVRSIEGAGPGPEGNITKLKLAEHMNE
MSMEG_0603|M.smegmatis_MC2_155 IEQNHAGQLIAEGHAAKVLNLRSTLLTLAGGDAMPNAAISKLLSMKTGQG
MAB_0580|M.abscessus_ATCC_1997 AQLDTLGAFVCSAQTGALLDLRTALRAIGGQDPGAASSVRKLVGVRHRQG
MLBr_00737|M.leprae_Br4923 SISTFQSIQFMLADMAMKVEAARLIVYAAAARAERGEPDLGFISAASKCF
. . :: . . :
Mflv_0441|M.gilvum_PYR-GCK LTELAFELAG--TAAVVEQMP----QLTRSYLGNRAMTIAGGTSEITRNT
Mvan_0226|M.vanbaalenii_PYR-1 LTELAFELAG--TAAVVGQTP----QLTRSYLGNRAMTIAGGTSEITRNT
Mb3087c|M.bovis_AF2122/97 GAAIAAALWGPEIALLDGPGR----VIGRTVMGARGMAIAGGTSEVTRNQ
Rv3061c|M.tuberculosis_H37Rv GAAIAAALWGPEIALLDGPGR----VIGRTVMGARGMAIAGGTSEVTRNQ
MMAR_1625|M.marinum_M GAAISAALLGPEIALLDGPGA----VIGRMVMGARGMAIAGGTSEVTRNQ
MUL_3411|M.ulcerans_Agy99 GAAISAALLGTEIALLDGPGA----VIGRMVMGARGMAIAGGTSEVTRNQ
MAV_3935|M.avium_104 GAAIMAALLGPEVALTDGPGA----LSGRLMMGARGMAIAGGTSEVTRNQ
TH_3278|M.thermoresistible__bu RAAIMAGLLGPELALLDGDTA----VAGLMVLGARGMAIAGGTSEVTRNQ
MSMEG_0603|M.smegmatis_MC2_155 YAEFAVSTFGTDAAIGDPQEEP--GRWAEYLLASRATTIYGGTTEVQLNI
MAB_0580|M.abscessus_ATCC_1997 LSEHILDYVDTHGAVES--------REMWLFMRDRCLSIAGGTTQILLSV
MLBr_00737|M.leprae_Br4923 ASDIAMEVTTDAVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVV
: : * **.::
Mflv_0441|M.gilvum_PYR-GCK IAERILGLPRDPLLR
Mvan_0226|M.vanbaalenii_PYR-1 IAERILGLPRDPLLR
Mb3087c|M.bovis_AF2122/97 IAERILGMPRDPLIS
Rv3061c|M.tuberculosis_H37Rv IAERILGMPRDPLIS
MMAR_1625|M.marinum_M IAERILGMPRDPLIN
MUL_3411|M.ulcerans_Agy99 IAERILGMPRDPLIN
MAV_3935|M.avium_104 IAERILGMPRDPLIN
TH_3278|M.thermoresistible__bu IAERILGLPRDPLIT
MSMEG_0603|M.smegmatis_MC2_155 IAERLLGLPRDP---
MAB_0580|M.abscessus_ATCC_1997 AGERLLGLPR-----
MLBr_00737|M.leprae_Br4923 MSRALLR--------
.. :*