For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TQDHIVTERPPATRNDTAGNATARTYEVRTLGCLMNAHDSERMAGLLEDAGYIKADPGAPADLVVFNTCA VRENADNKLYGSLAHLAPIKASRPGMQIAVGGCLAQKDRHIVLDRAPWVDVVFGTHNIGSLPVLLERSRH NQDAQVEILESLRTFPSALPAARDSAYAAWVSISVGCNNSCTFCIVPSLRGKEADRRPGDILAEVQALVE QGVLEVTLLGQNVNSYGVNFADPDPSTGEEPLPRDRGAFAQLLRACGRIEGLERIRFTSPHPAEFTDDVI LAMAETPAVCPHLHMPLQSGSDQILKAMRRSYRRDRYLGIIDKVRAAIPHAAITTDIIVGFPGETEHDFE QTLDVVQKARFTSAFTFQYSPRPGTPAADMPDQIPKNVVQQRFERLVALQERISLESNRSLVGTRQELLV VAGEGRKNATTARISGRARDGRLVHFRPDSADGSVRPGDLVDVEITDAAPHHLIADGPLLQHRRTPAGDA SERGQTPTTRGVGLGVPRITVAQAQIPSSGTHPDTACR*
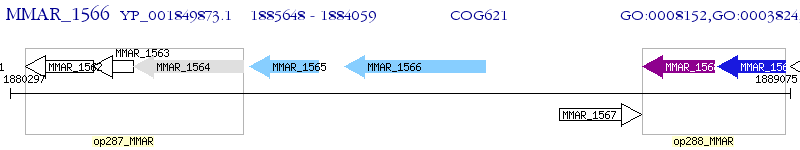
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1566 | miaB | - | 100% (529) | 2-methylthioadenine synthetase, MiaB |
| M. marinum M | MMAR_1979 | - | 0.0 | 74.09% (494) | hypothetical protein MMAR_1979 |
| M. marinum M | MMAR_1562 | - | 9e-05 | 30.15% (136) | hypothetical protein MMAR_1562 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2752c | - | 0.0 | 71.80% (500) | hypothetical protein Mb2752c |
| M. gilvum PYR-GCK | Mflv_3967 | - | 0.0 | 74.49% (486) | RNA modification protein |
| M. tuberculosis H37Rv | Rv2733c | - | 0.0 | 71.80% (500) | hypothetical protein Rv2733c |
| M. leprae Br4923 | MLBr_00989 | - | 0.0 | 68.75% (512) | (dimethylallyl)adenosine tRNA methylthiotransferase |
| M. abscessus ATCC 19977 | MAB_3048c | - | 0.0 | 72.89% (498) | putative tRNA-i(6)A37 modification enzyme MiaB |
| M. avium 104 | MAV_3625 | - | 0.0 | 76.28% (468) | hypothetical protein MAV_3625 |
| M. smegmatis MC2 155 | MSMEG_2729 | miaB | 0.0 | 74.31% (506) | tRNA-I(6)A37 thiotransferase enzyme MiaB |
| M. thermoresistible (build 8) | TH_1783 | - | 0.0 | 74.64% (489) | CONSERVED HYPOTHETICAL ALANINE, ARGININE-RICH PROTEIN |
| M. ulcerans Agy99 | MUL_3378 | - | 0.0 | 74.59% (488) | hypothetical protein MUL_3378 |
| M. vanbaalenii PYR-1 | Mvan_2429 | - | 0.0 | 74.59% (488) | RNA modification protein |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00989|M.leprae_Br4923 -------MVTRDAGMADDAAT---GSTRTYQVRTYGCQMNVHDSERLAGL
MAV_3625|M.avium_104 --------------------------MRTYQVRTYGCQMNVHDSERLAGL
Mb2752c|M.bovis_AF2122/97 MVAHDAAAGVTGEGAGPPVRR---APARTYQVRTYGCQMNVHDSERLAGL
Rv2733c|M.tuberculosis_H37Rv MVAHDAAAGVTGEGAGPPVRR---APARTYQVRTYGCQMNVHDSERLAGL
MUL_3378|M.ulcerans_Agy99 -MTSTVAHGAGSAGPADDVEP---MSARTYQVRTYGCQMNVHDSERMAGL
Mflv_3967|M.gilvum_PYR-GCK -------MLQQADGVSPDRSSCDTPAPRTFEVRTYGCQMNVHDSERLSGL
Mvan_2429|M.vanbaalenii_PYR-1 ---MTSTVTQQASGPSPDRS------ARTFEVRTYGCQMNVHDSERLAGL
MSMEG_2729|M.smegmatis_MC2_155 --MVTRDETAEAEGLRPAVGS---SRVRTYQVRTYGCQMNVHDSERLSGL
TH_1783|M.thermoresistible__bu --MVTRSRTGAGGASGAARER---SSARTYQIRTYGCQMNVHDSERLAGL
MAB_3048c|M.abscessus_ATCC_199 ------MTSTATSGES---------AVRTYQVRTYGCQMNVHDSERVSGL
MMAR_1566|M.marinum_M -MTQDHIVTERPPATRNDTAG--NATARTYEVRTLGCLMNAHDSERMAGL
**:::** ** **.*****::**
MLBr_00989|M.leprae_Br4923 LEAAGYQRAADEADVGDADVVVFNTCAVRENADNRLYGNLSHLAPRKRNN
MAV_3625|M.avium_104 LEAAGYRRAAEGAEV--ADVVVFNTCAVRENADNKLYGNLSHLAPRKRSN
Mb2752c|M.bovis_AF2122/97 LEAAGYRRATDGSEA---DVVVFNTCAVRENADNRLYGNLSHLAPRKRAN
Rv2733c|M.tuberculosis_H37Rv LEAAGYRRATDGSEA---DVVVFNTCAVRENADNRLYGNLSHLAPRKRAN
MUL_3378|M.ulcerans_Agy99 LEAAGYRRAAEGTDA---DVVVFNTCAVRENADNKLYGNLSHLAPRKRTS
Mflv_3967|M.gilvum_PYR-GCK LEQAGYQRAGAGVDA---DIVVFNTCAVRENADNKLYGNLSHLAPRKQAD
Mvan_2429|M.vanbaalenii_PYR-1 LEGAGYRRAGEGADA---DIVVFNTCAVRENADNKLYGNLSHLAPRKQSD
MSMEG_2729|M.smegmatis_MC2_155 LESAGYQRAAEGTDA---DIVVFNTCAVRENADNKLYGNLSHLAPRKQAD
TH_1783|M.thermoresistible__bu LEAAGYRRASDGADA---DVVVFNTCAVRENADNKLYGNLSHLAPRKQAD
MAB_3048c|M.abscessus_ATCC_199 LDAAGYVKAPEGTDA---DIVIFNTCAVRENADNKLYGNISHLAPRKAAN
MMAR_1566|M.marinum_M LEDAGYIKADPGAPA---DLVVFNTCAVRENADNKLYGSLAHLAPIKASR
*: *** :* . *:*:************:***.::**** *
MLBr_00989|M.leprae_Br4923 PDMQIAVGGCLAQKDKHTVLSKAPWVDIVFGTHNLGSLPTLLDRARHNKV
MAV_3625|M.avium_104 PQMQIAVGGCLAQKDREAVLRRAPWVDVVFGTHNIGSLPTLLERARHNKA
Mb2752c|M.bovis_AF2122/97 PDMQIAVGGCLAQKDRDAVLRRAPWVDVVFGTHNIGSLPTLLERARHNKV
Rv2733c|M.tuberculosis_H37Rv PDMQIAVGGCLAQKDRDAVLRRAPWVDVVFGTHNIGSLPTLLERARHNKV
MUL_3378|M.ulcerans_Agy99 PDMQIAVGGCLAQKDRDALLRKAPWVDVVFGTHNIGSLPALLDRARHNRV
Mflv_3967|M.gilvum_PYR-GCK PQMQIAVGGCLAQKDRDAVLRKAPWVDVVFGTHNIGSLPTLLDRARHNRV
Mvan_2429|M.vanbaalenii_PYR-1 PDMQIAVGGCLAQKDRDSVLRKAPWVDVVFGTHNIGSLPALLDRARHNRV
MSMEG_2729|M.smegmatis_MC2_155 PNMQIAVGGCLAQKDRDAVLRRAPWVDVVFGTHNIGSLPTLLERARHNRE
TH_1783|M.thermoresistible__bu PDLQIAVGGCLAQKDRDAVLRRAPWVDVVFGTHNIGSLPALLERARHNRT
MAB_3048c|M.abscessus_ATCC_199 PNMQIAVGGCLAQKDREGMLAKAPWVDVVFGTHNIGSLPALLERARHNNE
MMAR_1566|M.marinum_M PGMQIAVGGCLAQKDRHIVLDRAPWVDVVFGTHNIGSLPVLLERSRHNQD
* :************:. :* :*****:******:****.**:*:***.
MLBr_00989|M.leprae_Br4923 AQVEIVEALQHFPSSLPSARESDYAAWVSISVGCNNSCTFCIVPSLRGKE
MAV_3625|M.avium_104 AQVEIAEALQQFPSSLPSARESAYAAWVSISVGCNNSCTFCIVPSLRGKE
Mb2752c|M.bovis_AF2122/97 AQVEIAEALQQFPSSLPSSRESAYAAWVSISVGCNNSCTFCIVPSLRGRE
Rv2733c|M.tuberculosis_H37Rv AQVEIAEALQQFPSSLPSSRESAYAAWVSISVGCNNSCTFCIVPSLRGRE
MUL_3378|M.ulcerans_Agy99 AQVEIAEALQQFPSSLPSARESAYAAWVSISVGCNNTCTFCIVPSLRGKE
Mflv_3967|M.gilvum_PYR-GCK AQVEIAESLREFPSALPASRESAYAAWVSISVGCNNTCTFCIVPSLRGKE
Mvan_2429|M.vanbaalenii_PYR-1 AQVEIAEALQEFPSALPASRESSYAAWVSISVGCNNTCTFCIVPALRGRE
MSMEG_2729|M.smegmatis_MC2_155 AQVEIVEALQEFPSALPASRESAYAAWVSISVGCNNTCTFCIVPSLRGKE
TH_1783|M.thermoresistible__bu AQVEIAEALQEFPSNLPAARESAYAAWVSISVGCNNTCTFCIVPALRGKE
MAB_3048c|M.abscessus_ATCC_199 AQVEIVEALEHFPSALPATRESAYAAWVSISVGCNNTCTFCIVPSLRGKE
MMAR_1566|M.marinum_M AQVEILESLRTFPSALPAARDSAYAAWVSISVGCNNSCTFCIVPSLRGKE
***** *:*. *** **::*:* *************:*******:***:*
MLBr_00989|M.leprae_Br4923 VDRSPADILAEVEALVADGVLEVTLLGQNVNAYGVSFADP-------ALA
MAV_3625|M.avium_104 VDRSPDDILAEVRSLVADGVLEVTLLGQNVNAYGVSFADP-------ALP
Mb2752c|M.bovis_AF2122/97 VDRSPADILAEVRSLVNDGVLEVTLLGQNVNAYGVSFADP-------ALP
Rv2733c|M.tuberculosis_H37Rv VDRSPADILAEVRSLVNDGVLEVTLLGQNVNAYGVSFADP-------ALP
MUL_3378|M.ulcerans_Agy99 IDRSPADILAEVQSLVDTGVVEITLLGQNVNAYGVSFADP-------ALP
Mflv_3967|M.gilvum_PYR-GCK VDRRPGDVLAEVQALVDQGVLEITLLGQNVNAYGVSFADP-------TEA
Mvan_2429|M.vanbaalenii_PYR-1 VDRRPGDVLAEVQSLVDQGVLEITLLGQNVNAYGVSFADP-------TEP
MSMEG_2729|M.smegmatis_MC2_155 VDRRPGDILAEVQSLVDQGVLEITLLGQNVNAYGVSFADP-------ELP
TH_1783|M.thermoresistible__bu VDRRPDDILAEVRALVDQGVLEVTLLGQNVNAYGVSFADP-------DLP
MAB_3048c|M.abscessus_ATCC_199 IDRRPGDILGEVQALVDQGVLEVTLLGQNVNAYGVNFADP-------EIP
MMAR_1566|M.marinum_M ADRRPGDILAEVQALVEQGVLEVTLLGQNVNSYGVNFADPDPSTGEEPLP
** * *:*.**.:** **:*:********:***.**** .
MLBr_00989|M.leprae_Br4923 RNRGSFAELLRSCGSIDGLERVRFTSPHPAEFTYDVIEAMAQTPNVCPSL
MAV_3625|M.avium_104 RDRGAFARLLRACGEIDGLERVRFTSPHPAEFTDDVIEAMAQTPNVCPAL
Mb2752c|M.bovis_AF2122/97 RNRGAFAELLRACGDIDGLERVRFTSPHPAEFTDDVIEAMAQTRNVCPAL
Rv2733c|M.tuberculosis_H37Rv RNRGAFAELLRACGDIDGLERVRFTSPHPAEFTDDVIEAMAQTRNVCPAL
MUL_3378|M.ulcerans_Agy99 RNRGAFAELLRACGDIDGLERVRFTSPHPAEFTDDVIEAMAQTPNVCPAL
Mflv_3967|M.gilvum_PYR-GCK RDRGAFAKLLRACGRIDGLERVRFTSPHPAEFTDDVIEAMAQTSNVCPTL
Mvan_2429|M.vanbaalenii_PYR-1 RDRGAFAKLLRACGRIDGLERVRFTSPHPAEFTDDVIEAMAETPNVCPTL
MSMEG_2729|M.smegmatis_MC2_155 RDRGAFAKLLRACGGIDGLERVRFTSPHPAEFTDDVIEAMAATPNVCPTL
TH_1783|M.thermoresistible__bu RHRGAFADLLRACGEIDGLERVRFTSPHPAEFTDDVIEAMAETPNVCPAL
MAB_3048c|M.abscessus_ATCC_199 RDRGAFAELLRACGRIDGLERVRFTSPHPAEFTDDVIEAMAQTPNVCPQL
MMAR_1566|M.marinum_M RDRGAFAQLLRACGRIEGLERIRFTSPHPAEFTDDVILAMAETPAVCPHL
*.**:** ***:** *:****:*********** *** *** * *** *
MLBr_00989|M.leprae_Br4923 HMPLQSGSDRVLRAMRRSYRVERYLGIIGQVRAAMPHAAITTDLIVGFPG
MAV_3625|M.avium_104 HMPLQSGSDRVLRAMRRSYRAERYLGIIDRVRAAMPHAAITTDLIVGFPG
Mb2752c|M.bovis_AF2122/97 HMPLQSGSDRILRAMRRSYRAERYLGIIERVRAAIPHAAITTDLIVGFPG
Rv2733c|M.tuberculosis_H37Rv HMPLQSGSDRILRAMRRSYRAERYLGIIERVRAAIPHAAITTDLIVGFPG
MUL_3378|M.ulcerans_Agy99 HMPLQSGSDRVLRAMRRSYRAERYLGIIERVRAAMPHAAITTDLIVGFPG
Mflv_3967|M.gilvum_PYR-GCK HMPLQSGSDRILRAMRRSYRADRYLGIIDRVRTAIPHAAITTDLIVGFPG
Mvan_2429|M.vanbaalenii_PYR-1 HMPLQSGSDRILRAMRRSYRAERYLGIIDRVRAAIPHAAITTDLIVGFPG
MSMEG_2729|M.smegmatis_MC2_155 HMPLQSGSDRILKAMRRSYRAERFLGIIDKVRAAIPHAAITTDLIVGFPG
TH_1783|M.thermoresistible__bu HMPLQSGSDRILRAMRRSYRAERFLGILDKVRAAIPHAAITTDLIVGFPG
MAB_3048c|M.abscessus_ATCC_199 HMPLQSGSDRILKAMRRSYRSERFLSIIDKVRSAMPDAAITTDIIVGFPG
MMAR_1566|M.marinum_M HMPLQSGSDQILKAMRRSYRRDRYLGIIDKVRAAIPHAAITTDIIVGFPG
*********::*:******* :*:*.*: :**:*:*.******:******
MLBr_00989|M.leprae_Br4923 ETEEDFAATLDVVRQVRFTAAFTFQYSKRPGTPAAELGGQLPKAVVQERY
MAV_3625|M.avium_104 ETEEDFAATLDVVRRARFAAAFTFQYSKRPGTPAAELDGQIPKAVVQERY
Mb2752c|M.bovis_AF2122/97 ETEEDFAATLDVVRRARFAAAFTFQYSKRPGTPAAQLDGQLPKAVVQERY
Rv2733c|M.tuberculosis_H37Rv ETEEDFAATLDVVRRARFAAAFTFQYSKRPGTPAAQLDGQLPKAVVQERY
MUL_3378|M.ulcerans_Agy99 ETEQDFAATLDVVRQARFSAAFTFQYSKRPGTPAAELDGQLPKAVVQERY
Mflv_3967|M.gilvum_PYR-GCK ETEEDFQATLDVVEAARFSSAFTFQYSKRPGTPAAELEGQIPKAVVSERY
Mvan_2429|M.vanbaalenii_PYR-1 ETEEDFQATLDVVAASRFSSAFTFQYSKRPGTPAAELADQVPKAVVSERY
MSMEG_2729|M.smegmatis_MC2_155 ETEEDFQATLDVVEQARFAGAFTFQYSKRPGTPAADMPDQLPKSVVTERY
TH_1783|M.thermoresistible__bu ETEEDFQATLDVVEQARFAGAFTFQYSRRPGTPAADLPDQLPKAVVQERY
MAB_3048c|M.abscessus_ATCC_199 ETEEDFQQTLEVVRRARFSSAFTFQYSIRPGTPAAKLPDQLPKAVVQERY
MMAR_1566|M.marinum_M ETEHDFEQTLDVVQKARFTSAFTFQYSPRPGTPAADMPDQIPKNVVQQRF
***.** **:** **:.******* *******.: .*:** ** :*:
MLBr_00989|M.leprae_Br4923 ERLVELQEQICMEENRVLIGRIVELLVTTGEGRKDARTARRTGRARDGRL
MAV_3625|M.avium_104 ERLVELQESISLQGNQALVGQTVELLVATGEGRKDSATARMSGRARDGRL
Mb2752c|M.bovis_AF2122/97 ERLIALQEQISLEANRALVGQAVEVLVATGEGRKDTVTARMSGRARDGRL
Rv2733c|M.tuberculosis_H37Rv ERLIALQEQISLEANRALVGQAVEVLVATGEGRKDTVTARMSGRARDGRL
MUL_3378|M.ulcerans_Agy99 ERLVELQEQISLEGNRAIVGQRVELLVATGEGRKDTLTARMSGRARDGRL
Mflv_3967|M.gilvum_PYR-GCK QRLIELQERISWEENRAQIGREVELLVATGEGRKDAATARMSGRARDGRL
Mvan_2429|M.vanbaalenii_PYR-1 QRLIELQERISLEENTAQIGRRVELLVATGEGRKDAATARMSGRARDGRL
MSMEG_2729|M.smegmatis_MC2_155 QRLIELQERISLEQNREQVGRAVELLVATGEGRKDASTARMSGRARDGRL
TH_1783|M.thermoresistible__bu ERLIALQDRIALEENTAQIGRAVELLVATGEGRKDASTARMSGRARDGRL
MAB_3048c|M.abscessus_ATCC_199 DRLIALQESVTLEENQKQIGRMIEVLIATGEGRKDGETARMSGRARDGRL
MMAR_1566|M.marinum_M ERLVALQERISLESNRSLVGTRQELLVVAGEGRKNATTARISGRARDGRL
:**: **: : : * :* *:*:.:*****: *** :********
MLBr_00989|M.leprae_Br4923 VHFALDTPWEAQVRPGDIITVMVTEAAPHHLIADAGILTHRRTRAGDAHA
MAV_3625|M.avium_104 VHFAADD----RVRPGDLVTTVITGAAPHHLIADAGILSHRRTRAGDAHA
Mb2752c|M.bovis_AF2122/97 VHFTAGQP---RVRPGDVITTKVTEAAPHHLIADAGVLTHRRTRAGDAHT
Rv2733c|M.tuberculosis_H37Rv VHFTAGQP---RVRPGDVITTKVTEAAPHHLIADAGVLTHRRTRAGDAHT
MUL_3378|M.ulcerans_Agy99 VHFRAGDG---PVRPGDIVTVEVTDAAPHHLIADGGILAHRRTRAGDAHA
Mflv_3967|M.gilvum_PYR-GCK VHFAPGALG-ADIRPGDIVVTTVTGAAPHHLIADAGPAEHRRTRAGDAHA
Mvan_2429|M.vanbaalenii_PYR-1 VHFAPGAVG-SEVRPGDVVVTTVTGAAPHHLIADAGLIEHRRTRAGDAHA
MSMEG_2729|M.smegmatis_MC2_155 VHFLPGDN---EIRPGDIVTTTVTGAAPHHLIADAPIIERRRTRAGDAHA
TH_1783|M.thermoresistible__bu VHFTPGGL---DIRPGDIVTTTVTAAAPHYLIADAEPAGHRRTRAGDAHA
MAB_3048c|M.abscessus_ATCC_199 VHFRPQGHVDGALRPGDIITVDVTGAAPHHLIADDGVRSHRRTRAGDAHE
MMAR_1566|M.marinum_M VHFRPDSADG-SVRPGDLVDVEITDAAPHHLIADGPLLQHRRTPAGDASE
*** :****:: . :* ****:**** :*** ****
MLBr_00989|M.leprae_Br4923 AR--------QCPRS-----------------------------------
MAV_3625|M.avium_104 PAGARAASVSACPPSGRRRPGPTRWMCLMNDDRNHDDPRLGALRTEIEAA
Mb2752c|M.bovis_AF2122/97 AG--------QPGRA-----------------------------------
Rv2733c|M.tuberculosis_H37Rv AG--------QPGRA-----------------------------------
MUL_3378|M.ulcerans_Agy99 DG--------QTVRG-----------------------------------
Mflv_3967|M.gilvum_PYR-GCK EG--------RTPKTG----------------------------------
Mvan_2429|M.vanbaalenii_PYR-1 AG--------QKPRTG----------------------------------
MSMEG_2729|M.smegmatis_MC2_155 AG--------QKPRTG----------------------------------
TH_1783|M.thermoresistible__bu AG--------LRPKT-----------------------------------
MAB_3048c|M.abscessus_ATCC_199 AG--------KKPSTPG---------------------------------
MMAR_1566|M.marinum_M RG--------QTPTTRG---------------------------------
MLBr_00989|M.leprae_Br4923 -------IGLG----------------------------QIPLGMPSVGQ
MAV_3625|M.avium_104 ERRVAGGIDPGARGFVVSILVFVLLGSFILPHTGDVRGWDVLFGTHDAGA
Mb2752c|M.bovis_AF2122/97 ----------------------------------------VGLGMPGVG-
Rv2733c|M.tuberculosis_H37Rv ----------------------------------------VGLGMPGVG-
MUL_3378|M.ulcerans_Agy99 ----------------------------------------IGLGMPGIGR
Mflv_3967|M.gilvum_PYR-GCK ----------------------------------------VGLGMPGIGA
Mvan_2429|M.vanbaalenii_PYR-1 ----------------------------------------VGLGMPAVGA
MSMEG_2729|M.smegmatis_MC2_155 ----------------------------------------VGLGMPRIGA
TH_1783|M.thermoresistible__bu ----------------------------------------VGLGLPRIGG
MAB_3048c|M.abscessus_ATCC_199 ----------------------------------------IGLGMPAIGA
MMAR_1566|M.marinum_M ----------------------------------------VGLGVPRITV
: :*
MLBr_00989|M.leprae_Br4923 HSASIGS------------------------------------AGCAR--
MAV_3625|M.avium_104 AAVALPSRVFGWLALVFGVGFSTLALVTRRWALAWIALAGTAIAGAAGML
Mb2752c|M.bovis_AF2122/97 --LPVSAAKP---------------------------------GGCR---
Rv2733c|M.tuberculosis_H37Rv --LPVSAAKP---------------------------------GGCR---
MUL_3378|M.ulcerans_Agy99 PVVPVAAEATSC----------------------------GSAGGCGSAD
Mflv_3967|M.gilvum_PYR-GCK PEPAPV------------------------------------TQGCAL--
Mvan_2429|M.vanbaalenii_PYR-1 PDPLPA------------------------------------TTGCAR--
MSMEG_2729|M.smegmatis_MC2_155 PAPSAT------------------------------------AEGCGC--
TH_1783|M.thermoresistible__bu PKPDAEP-----------------------------------TRGCGC--
MAB_3048c|M.abscessus_ATCC_199 PKTERIE-----------------------------------VAGCGL--
MMAR_1566|M.marinum_M AQAQIPSSGTHP------------------------------DTACR---
..
MLBr_00989|M.leprae_Br4923 --------------------------------------------------
MAV_3625|M.avium_104 AIWSRQTVPAGHPGPGWGLIVAWITVLVLIYQWARVVWSRTIVQLAAEEQ
Mb2752c|M.bovis_AF2122/97 --------------------------------------------------
Rv2733c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_3378|M.ulcerans_Agy99 GAGSSAGDPQ----------------------------------------
Mflv_3967|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2429|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2729|M.smegmatis_MC2_155 --------------------------------------------------
TH_1783|M.thermoresistible__bu --------------------------------------------------
MAB_3048c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1566|M.marinum_M --------------------------------------------------
MLBr_00989|M.leprae_Br4923 ---------------------------
MAV_3625|M.avium_104 RRRVAAQQQSTTLLDDLPKPEDPAAGT
Mb2752c|M.bovis_AF2122/97 ---------------------------
Rv2733c|M.tuberculosis_H37Rv ---------------------------
MUL_3378|M.ulcerans_Agy99 ---------------------------
Mflv_3967|M.gilvum_PYR-GCK ---------------------------
Mvan_2429|M.vanbaalenii_PYR-1 ---------------------------
MSMEG_2729|M.smegmatis_MC2_155 ---------------------------
TH_1783|M.thermoresistible__bu ---------------------------
MAB_3048c|M.abscessus_ATCC_199 ---------------------------
MMAR_1566|M.marinum_M ---------------------------