For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLEGPLADRDGWSAVGECPIEKTMAVVGTKSAMLIMREAYYGTTRFDDFARRVGITKAATSARLAELVSL KLLARRPYREPGQRSRDEYVLTEAGLDFMPVVFSLFQWGKRHLPGGDRLQLTHLGCGAQAQVEIRCAKGH LVPADELGMRLVRSP
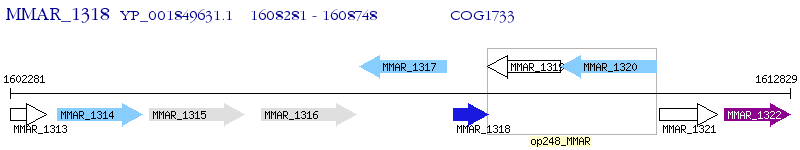
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1318 | - | - | 100% (155) | transcriptional regulatory protein |
| M. marinum M | MMAR_1555 | - | 3e-15 | 35.83% (120) | transcriptional regulatory protein |
| M. marinum M | MMAR_1626 | - | 1e-07 | 31.63% (98) | transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3122 | - | 3e-17 | 37.82% (119) | putative transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_3781 | - | 1e-64 | 75.82% (153) | HxlR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3095 | - | 3e-17 | 37.82% (119) | putative transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4427c | - | 6e-30 | 42.11% (152) | putative transcriptional regulator |
| M. avium 104 | MAV_4183 | - | 3e-74 | 84.97% (153) | putative transcriptional regulator family protein |
| M. smegmatis MC2 155 | MSMEG_3013 | - | 3e-61 | 70.13% (154) | putative transcriptional regulator family protein |
| M. thermoresistible (build 8) | TH_2405 | - | 9e-05 | 29.79% (94) | putative transcriptional regulator |
| M. ulcerans Agy99 | MUL_2562 | - | 4e-87 | 99.35% (155) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_2618 | - | 8e-66 | 75.66% (152) | HxlR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3122|M.bovis_AF2122/97 -------MAVSDLSHRFEGESVGRALELVGERWTLLILREAFFGVRRFGQ
Rv3095|M.tuberculosis_H37Rv -------MAVSDLSHRFEGESVGRALELVGERWTLLILREAFFGVRRFGQ
MMAR_1318|M.marinum_M --MLEGPLADRDGWSAVGECPIEKTMAVVGTKSAMLIMREAYYGTTRFDD
MUL_2562|M.ulcerans_Agy99 --MLEGPLADRDGWSAVGECPIEKTMAVVGTKSAMLIMREAYYGTTRFDD
MAV_4183|M.avium_104 MTLLQGPLADRDAWSAVGECAIEKTMAVVGTKSAMLIMREAYYGTTRFDD
Mflv_3781|M.gilvum_PYR-GCK MTVLQGPLADRSAWSAVGRCPIEKTMALVGTRSAMLLMREAYYGTTRFDD
Mvan_2618|M.vanbaalenii_PYR-1 MPVLQGPLVDRDAWSAVGECPIEKTMAVAGQKSAMLIMREAYYGTTRFDD
MSMEG_3013|M.smegmatis_MC2_155 MAMLQGALADRDAWSPVGRCPIEKAMGLVGTKSAMLIMREAYYGTTRFED
MAB_4427c|M.abscessus_ATCC_199 -MGFERPLRDRSRWN-TDACSIGKALDLLSTKTAFLIVRECFYGSTRFDE
TH_2405|M.thermoresistible__bu -------MASR---TYGQHCALAKSLDLVGDRWTLLIVRELLDGPRRYSD
: .: ::: : . : ::*::** * *: :
Mb3122|M.bovis_AF2122/97 LARNLG-IPRPTLSSRLRMLVEVGLFDRVPYS--SDPERHEYRLTEAGRD
Rv3095|M.tuberculosis_H37Rv LARNLG-IPRPTLSSRLRMLVEVGLFDRVPYS--SDPERHEYRLTEAGRD
MMAR_1318|M.marinum_M FARRVG-ITKAATSARLAELVSLKLLARRPYREPGQRSRDEYVLTEAGLD
MUL_2562|M.ulcerans_Agy99 FARRVG-ITKAATSARLAELVSLKLLARRPYREPGQRSRDEYVLTEAGLD
MAV_4183|M.avium_104 FARRVG-ITKAATSARLAELVELGLLKRRPYREPGQRSRDEYVLTQAGID
Mflv_3781|M.gilvum_PYR-GCK FSRRVG-ITKAATSARLAELTEAGLLTKRPYREPGQRVRDEYVLTEAGTE
Mvan_2618|M.vanbaalenii_PYR-1 FAKRVG-ITKAATSARLSELVEAGLLTKQPYREPGQRARDEYVLTQAGVD
MSMEG_3013|M.smegmatis_MC2_155 FWRRVG-VTKAAASARLSDLVEAGLLERRPYQEPGQRVRDEYVLTDAGRD
MAB_4427c|M.abscessus_ATCC_199 FAERIG-VSAPAVSRALKQLEAAGVIEKVPYREPGQRERDGYVLTRMGED
TH_2405|M.thermoresistible__bu LLTALRPIATDMLATRLRHLEEHHLVRRRELPKPASGT--VYELTADGAA
: : :. : * * :. : .. * ** *
Mb3122|M.bovis_AF2122/97 LFAAIVVLMQWGDEYLPRPEGPPIKLRHHTCGEHADPRLICTHCGEEITA
Rv3095|M.tuberculosis_H37Rv LFAAIVVLMQWGDEYLPRPEGPPIKLRHHTCGEHADPRLICTHCGEEITA
MMAR_1318|M.marinum_M FMPVVFSLFQWGKRHLPGGDR--LQLTHLGCGAQAQVEIRCAKG------
MUL_2562|M.ulcerans_Agy99 FMPVVFSLFQWGKRHLPGGDR--LQLTHLGCGAQAQVEIRCAKG------
MAV_4183|M.avium_104 FMPVVWAMFEWGRRHLPGRHR--LQLTHLGCGAEVGVEIRCAEG------
Mflv_3781|M.gilvum_PYR-GCK FMPVVWAMFEWGRKHLDDTS---LRLTHLGCGADASVDIVCDKG------
Mvan_2618|M.vanbaalenii_PYR-1 FMPVVWAMFEWGRSHLPNNGR--LRLTHVPCGADAHVEIVCDNG------
MSMEG_3013|M.smegmatis_MC2_155 FMPVVWALFEWGRRHIPNDSP--LRLAHDGCGAYATVEVRCDDG------
MAB_4427c|M.abscessus_ATCC_199 LLPVLLSLLKWGDDYLQNGHKP-LTLIDKKTGGEIGVQVTSRPG------
TH_2405|M.thermoresistible__bu LEAVVHAFIRWGRSLLETRRAG-DVVRPHWLARAVRAHVRPDRTGPPLSV
: ..: ::.** : : . :
Mb3122|M.bovis_AF2122/97 RNVTPEPGPGFKAKLASS--------------------------------
Rv3095|M.tuberculosis_H37Rv RNVTPEPGPGFKAKLASS--------------------------------
MMAR_1318|M.marinum_M -HLVPADELGMRLVRSP---------------------------------
MUL_2562|M.ulcerans_Agy99 -HLMPADELGMRLVRSP---------------------------------
MAV_4183|M.avium_104 -HLVPPDELGMRLAKKSG--------------------------------
Mflv_3781|M.gilvum_PYR-GCK -HSVPPDELGMRLVRRNRSADGG---------------------------
Mvan_2618|M.vanbaalenii_PYR-1 -HSVPLEELGMRLVPQKRPESG----------------------------
MSMEG_3013|M.smegmatis_MC2_155 -HVVPPDELVIRVARA----------------------------------
MAB_4427c|M.abscessus_ATCC_199 -QAVSADGIEVRVARR----------------------------------
TH_2405|M.thermoresistible__bu RLVTPEGAAAVRISADAVEDLDDDTAVDVTLTGAAEVLAAALHPDRAAEL
. .:
Mb3122|M.bovis_AF2122/97 ----------------------------------
Rv3095|M.tuberculosis_H37Rv ----------------------------------
MMAR_1318|M.marinum_M ----------------------------------
MUL_2562|M.ulcerans_Agy99 ----------------------------------
MAV_4183|M.avium_104 ----------------------------------
Mflv_3781|M.gilvum_PYR-GCK ----------------------------------
Mvan_2618|M.vanbaalenii_PYR-1 ----------------------------------
MSMEG_3013|M.smegmatis_MC2_155 ----------------------------------
MAB_4427c|M.abscessus_ATCC_199 ----------------------------------
TH_2405|M.thermoresistible__bu VAQGRLRVDGRADALRRLAEVVTAPAGRAVPDRS