For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVKKNAALSAQIIDTKRPNIVGADKHPGWHALRKIASRITTPLLPDDYLQLANPLWSARELRGRILEVRR ETEDSATLVIKPGWGFSFDYQPGQYIGIGLLVDGRWRWRSYSLTSSPVGGKSGPGSARTVTITVKAMPEG FLSTHLVAGVAPGTIVRLVAPQGNFVLPDPAPPSMLFLTAGSGITPVMSMLRTLLRRNQITDITHLHSVP TEADVMFGDELAGVAAQHPGYRLSVRATRSQGRLDLSRIGEQVPDWRERQTWACGPESMLNQAEKVWSAA GIGDRLHLERFAVSKAAPAGAGGTVTFARSGRSVDVDAATSVMDAGERAGVQMPFGCRMGICQSCVVDLV QGHVRDLRTGQEHDPGSRVQTCVTAASGDCVIDV
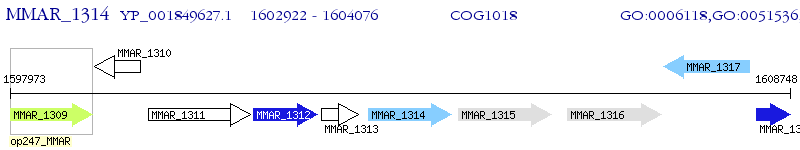
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1314 | - | - | 100% (384) | oxidoreductase |
| M. marinum M | MMAR_0335 | - | 3e-36 | 31.58% (361) | flavodoxin oxidoreductase |
| M. marinum M | MMAR_5066 | hmp | 9e-22 | 26.63% (368) | flavodoxin reductase Hmp |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3259c | - | 0.0 | 83.85% (384) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4699 | - | 1e-170 | 74.94% (387) | ferredoxin |
| M. tuberculosis H37Rv | Rv3230c | - | 0.0 | 84.11% (384) | oxidoreductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3549c | - | 1e-153 | 68.90% (373) | putative oxidoreductase |
| M. avium 104 | MAV_4193 | - | 0.0 | 80.73% (384) | 2Fe-2S iron-sulfur cluster binding domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_1885 | - | 1e-178 | 75.26% (384) | 2Fe-2S iron-sulfur cluster binding domain-containing protein |
| M. thermoresistible (build 8) | TH_2872 | - | 1e-168 | 73.18% (384) | PUTATIVE 2Fe-2S iron-sulfur cluster binding domain protein |
| M. ulcerans Agy99 | MUL_2566 | - | 0.0 | 99.22% (384) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_1767 | - | 1e-174 | 76.30% (384) | ferredoxin |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1314|M.marinum_M ----MVKKNAALSAQIIDTKRPNIVGADKHPGWHALRKIASRITTPLLPD
MUL_2566|M.ulcerans_Agy99 ----MVKKNAALSAQIIDTKRPSIVGADKHPGWHALRKIASRITTPLLPD
Mb3259c|M.bovis_AF2122/97 ----MSKKHTTLNASIIDTRRPTVAGADRHPGWHALRKIAARITTPLLPD
Rv3230c|M.tuberculosis_H37Rv ----MSKKHTTLNASIIDTRRPTVAGADRHPGWHALRKIAARITTPLLPD
MAV_4193|M.avium_104 MSNTMSKTTSPITANLGDTKRPVVAGAERHPGWHALRKLAARITTPLLPD
Mflv_4699|M.gilvum_PYR-GCK ----MAKSTIKNTAKVADTVRPAVAGAKERPLWHALRTVAGRITTPLLPD
Mvan_1767|M.vanbaalenii_PYR-1 ----MAKNTIKNTAKVADTVRPTIAGAKENPGWHLLRTIAGRITTPLLPD
MSMEG_1885|M.smegmatis_MC2_155 ----MAKNHIKISANVADTVRPTVAGADERPGWHALRRLAARITTPLLPD
TH_2872|M.thermoresistible__bu ----MAKSEIKYSARVVDTVRPQVAGVENRPVLHRLRRFAERITTPLLPD
MAB_3549c|M.abscessus_ATCC_199 ----------MKLASWLDASAPGAP-KSRHAAVNILRGLASRITTPLLPD
* *: * .... : ** .* *********
MMAR_1314|M.marinum_M DYLQLANPLWSARELRGRILEVRRETEDSATLVIKPGWGFSFDYQPGQYI
MUL_2566|M.ulcerans_Agy99 DYLQLANPLWSARELRGRILEVRRETEDSATLVIKPGWGFSFDYQPGQYI
Mb3259c|M.bovis_AF2122/97 DYLHLANPLWSARELRGRILGVRRETEDSATLFIKPGWGFSFDYQPGQYI
Rv3230c|M.tuberculosis_H37Rv DYLHLANPLWSARELRGRILGVRRETEDSATLFIKPGWGFSFDYQPGQYI
MAV_4193|M.avium_104 DYLHLANPLWSARELRGRVVDVRRETEDSATLVIKPGWGFNFDYQPGQYI
Mflv_4699|M.gilvum_PYR-GCK DYLQLANPLWSARELRGRVLEVRRETVDSATLVIKPGWGFSFDYEAGQYI
Mvan_1767|M.vanbaalenii_PYR-1 DYLKLANPLWSARELRGRVLEVRRETEDSATLVIKPGWGFSFDYEPGQYI
MSMEG_1885|M.smegmatis_MC2_155 DYLQLANPLWSARELRGRVLEVRRETEDSATLVIKPGWGFSFDYQPGQYI
TH_2872|M.thermoresistible__bu DYLHLINPLWSARELRGRVEDVRRETEDSATLVIKPGWGFSFDYQPGQYI
MAB_3549c|M.abscessus_ATCC_199 DYLSLANPLWSARELRGRVVSVRKETSDSATLEIKPGWGFHFDYHPGQYV
*** * ************: **:** ***** ******* ***..***:
MMAR_1314|M.marinum_M GIGLLVDGRWRWRSYSLTSSPVGGK--------SGPGSARTVTITVKAMP
MUL_2566|M.ulcerans_Agy99 GIGLLVDGRWRWRSYSLTSSPVGGK--------SGPGSARTVTITVKAMP
Mb3259c|M.bovis_AF2122/97 GIGLLVDGCWRWRSYSLTSSPAAS------------GSARMVTVTVKAMP
Rv3230c|M.tuberculosis_H37Rv GIGLLVDGRWRWRSYSLTSSPAAS------------GSARMVTVTVKAMP
MAV_4193|M.avium_104 GIGLLIDGRWRWRSYSLTSSPATS------------ASARTVTITVKAMP
Mflv_4699|M.gilvum_PYR-GCK GIGLLVEGRWRWRSYSLTSPPAER---------PARS-ARTITITVKAMP
Mvan_1767|M.vanbaalenii_PYR-1 GIGLLVEGRWRWRSYSLTSSPVT-----------SKG-SRTITITVKAMP
MSMEG_1885|M.smegmatis_MC2_155 GIGVFLDGRWRWRSYSLTSSPVTA---------HPKGRARTITITVKAMP
TH_2872|M.thermoresistible__bu GIGLPVDGRWRWRSYSLTSPAVTS---------GRGG--RTITITVKAMP
MAB_3549c|M.abscessus_ATCC_199 GIGVLIDGRWRWRSYSLTSAPAMSGSREERRALHAGGSARTITITVKAMP
***: ::* **********... . * :*:******
MMAR_1314|M.marinum_M EGFLSTHLVAGVAPGTIVRLVAPQGNFVLPDPAPPSMLFLTAGSGITPVM
MUL_2566|M.ulcerans_Agy99 EGFLSTHLVAGVAPGTIVRLVAPQGNFVLPDPAPPSILFLTAGSGITPVM
Mb3259c|M.bovis_AF2122/97 EGFLSTHLVAGVKPGTIVRLAAPQGNFVLPDPAPPLILFLTAGSGITPVM
Rv3230c|M.tuberculosis_H37Rv EGFLSTHLVAGVKPGTIVRLAAPQGNFVLPDPAPPLILFLTAGSGITPVM
MAV_4193|M.avium_104 EGFLSSHLVAGVEPGTVVRLAAPQGNFVLPDPAPASILFLTAGSGITPVM
Mflv_4699|M.gilvum_PYR-GCK EGFLSTHLVGGVEPGAIVRLAAPQGNFVMPDPAPASVLFLTAGSGITPVM
Mvan_1767|M.vanbaalenii_PYR-1 EGFLSTHLVGGVAPGTIVRLAAPQGNFVMPDPAPASVLFLTAGSGITPVM
MSMEG_1885|M.smegmatis_MC2_155 EGFLSTHLVGGLEPGTVVRLAAPQGNFVMPDPAPASVLFLTAGSGITPVM
TH_2872|M.thermoresistible__bu EGFLSTHLVEGVKPGTIVRLAAPQGNFVMPNPAPPQVLFLTAGSGITPIM
MAB_3549c|M.abscessus_ATCC_199 EGFLSTHLVNGLEPGTIVRLAAPQGNFVLDDPAPPAILFLTAGSGITPIM
*****:*** *: **::***.*******: :***. :***********:*
MMAR_1314|M.marinum_M SMLRTLLRR-----NQITDITHLHSVPTEADVMFGDELAGVAAQHPGYRL
MUL_2566|M.ulcerans_Agy99 SMLRTLLRR-----NQITDITHLHSVPTEADVMFGDELAGVAAQHPGYRL
Mb3259c|M.bovis_AF2122/97 SMLRTLVRR-----NQITDVVHLHSAPTAADVMFGAELAALAADHPGYRL
Rv3230c|M.tuberculosis_H37Rv SMLRTLVRR-----NQITDVVHLHSAPTAADVMFGAELAALAADHPGYRL
MAV_4193|M.avium_104 SMLRTLVRR-----NQIGDIVHLHSAPTEADVMFRGELAALANEHPGYRL
Mflv_4699|M.gilvum_PYR-GCK SMLRTLVRRDAI--GAPGHIVHVHSAPTEADIMFAAELAELARSHEGYRL
Mvan_1767|M.vanbaalenii_PYR-1 SMLRTLVRR-----GQLGDVAHVHSSPTEDDVMFAAELSELADRHEGYRL
MSMEG_1885|M.smegmatis_MC2_155 SMLRTLVRR-----DQITDIVHLHSAPTESDVLFGSELTALQGTHPGYRL
TH_2872|M.thermoresistible__bu SMLRTLERR-----DQITDIVHIHSVPTPSDAMFSGELADLQRRHPSYRF
MAB_3549c|M.abscessus_ATCC_199 SMLRTLVRRSAAAGLPLPDVVHLHSAPTAGDVMFAGELAAFSEAHPSYHY
****** ** .:.*:** ** * :* **: . * .*:
MMAR_1314|M.marinum_M SVRATRSQGRLDLSRIGEQVPDWRERQTWACGPESMLNQAEKVWSAAGIG
MUL_2566|M.ulcerans_Agy99 SVRATRSQGRLDLSRIGEQVPDWRERQTWACGPESMLNQAEKVWSAAGIG
Mb3259c|M.bovis_AF2122/97 SVRETRAQGRLDLTRIGQQVPDWRERQTWACGPEGVLNQADKVWSSAGAS
Rv3230c|M.tuberculosis_H37Rv SVRETRAQGRLDLTRIGQQVPDWRERQTWACGPEGVLNQADKVWSSAGAS
MAV_4193|M.avium_104 QLRETRTRGRLDLSLLDREVPDWRERQTWACGPEGMLNAAERVWSAAGIG
Mflv_4699|M.gilvum_PYR-GCK ELRATRTQGRLDVGRLDDVVPDWRDRQTWACGPEAMLEDAERTWKSAGVG
Mvan_1767|M.vanbaalenii_PYR-1 RVRATRTEGRLDLDRLDEVVPDWRERQTWACGPEAMLEAAERTWKAAGIA
MSMEG_1885|M.smegmatis_MC2_155 RIRSTRTEGRLDLERIADEVPDWRERQVWACGPEGMLADAERVWKAAGVG
TH_2872|M.thermoresistible__bu RLRTTRTEGRLDLSRLDDEVGDWRSRQTWACGPEGLLTAAERVWLGAGIG
MAB_3549c|M.abscessus_ATCC_199 RLRTTKTQGRLDFSSLAAEVPDWARRQTWACGPEGMLDDAQRTWADAGLA
:* *::.****. : * ** **.******.:* *::.* ** .
MMAR_1314|M.marinum_M DRLHLERFAVSKAAPAGAGGTVTFARSGRSVDVDAATSVMDAGERAGVQM
MUL_2566|M.ulcerans_Agy99 DRLHLERFAVSKAAPAGAGGTVTFARSGRSVDVDAATSVMDAGERAGVQM
Mb3259c|M.bovis_AF2122/97 DRLHLERFAVSKTAPAGAGGTVTFARSGKSVAADAATSLMDAGEGAGVQL
Rv3230c|M.tuberculosis_H37Rv DRLHLERFAVSKTAPAGAGGTVTFARSGKSVAADAATSLMDAGEGAGVQL
MAV_4193|M.avium_104 EQLHLERFAVSKAAPAGAGGMVTFARSGRSVDADAATSLMDAGEAAGVQM
Mflv_4699|M.gilvum_PYR-GCK ERLHQERFAVSRAPVHGSGGTVTFARSDRTVTVDAATSLMDAGEDAGVQM
Mvan_1767|M.vanbaalenii_PYR-1 DRLHLERFAVARA-AHGSGGTVEFSRSGKAATVDGATSLMDAGEAAGIQM
MSMEG_1885|M.smegmatis_MC2_155 ENLHLERFAVKRAAPHGTGGNVTFERTGKTVHADAAKSLMEAGEEVGIRM
TH_2872|M.thermoresistible__bu DRLHLERFAVARVANPGGGGTIEFARSGKSTVVDAATSLMDAGEQAGVQM
MAB_3549c|M.abscessus_ATCC_199 ELLHLERFAVSRAAPHGQGGLVTFARSGKQRAADAATSLMEAGEEAGVQM
: ** ***** :. * ** : * *:.: .*.*.*:*:*** .*:::
MMAR_1314|M.marinum_M PFGCRMGICQSCVVDLVQGHVRDLRTGQEHDPGSRVQTCVTAASGDCVID
MUL_2566|M.ulcerans_Agy99 PFGCRMGICQSCVVDLVQGHVRDLRTGQEHDPGSRVQTCVTAASGDCAID
Mb3259c|M.bovis_AF2122/97 PFGCRMGICQSCVVDLVEGHVRDLRTGQRHEPGTRVQTCVSAASGDCVLD
Rv3230c|M.tuberculosis_H37Rv PFGCRMGICQSCVVDLVEGHVRDLRTGQRHEPGTRVQTCVSAASGDCVLD
MAV_4193|M.avium_104 PFGCRMGICQSCVVGLVEGHVRDLRTGAEHEPGTRVQTCVSAASGDCVLD
Mflv_4699|M.gilvum_PYR-GCK PFGCRMGICQSCVVGLVEGHVRDLRTGIEHEPGSRVQTCVTAASGDCVLD
Mvan_1767|M.vanbaalenii_PYR-1 PFGCRMGICQSCVVGLLDGHVRDLRTGVEHEPGTRVQTCVSAASGDCVLD
MSMEG_1885|M.smegmatis_MC2_155 PFGCRMGICQSCVVGLVDGHVRDLRTGVEHDAGSRIQTCVSAASGDCVLD
TH_2872|M.thermoresistible__bu PFGCRMGICQSCVIGLVDGHVRDLRTGEVHDPGTRVQTCVSAAAGDCVLD
MAB_3549c|M.abscessus_ATCC_199 PFGCRMGICQTCVVSLVQGHARDLRTGAEYEPGTRIQTCVSAASGDCVLD
**********:**:.*::**.****** ::.*:*:****:**:***.:*
MMAR_1314|M.marinum_M V
MUL_2566|M.ulcerans_Agy99 V
Mb3259c|M.bovis_AF2122/97 I
Rv3230c|M.tuberculosis_H37Rv I
MAV_4193|M.avium_104 I
Mflv_4699|M.gilvum_PYR-GCK V
Mvan_1767|M.vanbaalenii_PYR-1 V
MSMEG_1885|M.smegmatis_MC2_155 V
TH_2872|M.thermoresistible__bu I
MAB_3549c|M.abscessus_ATCC_199 V
: