For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MFTQTLTQTLAKRVLGSGLVDLLTGPHGIDRYTELVEPTWTLGEARAKLIDVRRTTPRSVTLTLAPNDTF RSAMAAGTGPMAGQYVNLTVDIDGRRHTRCYSPANAEGSETLELTIGRHDGGLVSTYLCDHARRGMVVGL AGVGGDFVLPQRRPRRILFVSGGSGITPVLAMLRTLVAQHHLHIHGAEIAFIHYARNPAEACYRAELAAL PGVRVLHGYTQSGGGDLVGRFGPDHLAAAMDAPDAVFVCGPTTLVDAVRENCENVFTESFVPAPIEAPAQ PSGGRVRFADSGIDVVDDGRSLLEQAESAGLAPENGCRMGICHTCTRRKTSGTVRNLVTGAVSVAPDEDV QVCVSVPVGDVDLSL
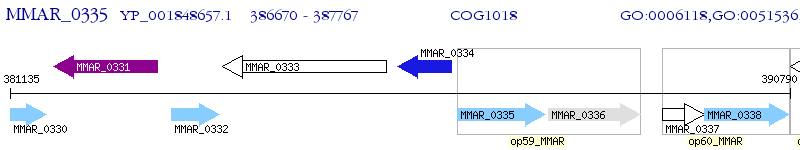
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0335 | - | - | 100% (365) | flavodoxin oxidoreductase |
| M. marinum M | MMAR_1314 | - | 3e-36 | 31.58% (361) | oxidoreductase |
| M. marinum M | MMAR_2865 | - | 2e-14 | 35.68% (199) | oxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3259c | - | 4e-38 | 33.70% (362) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4815 | - | 1e-124 | 63.24% (370) | oxidoreductase FAD-binding subunit |
| M. tuberculosis H37Rv | Rv3230c | - | 5e-39 | 33.98% (362) | oxidoreductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1588c | - | 3e-71 | 46.28% (363) | putative oxidoreductase |
| M. avium 104 | MAV_5168 | - | 1e-169 | 79.57% (372) | oxidoreductase FAD-binding subunit |
| M. smegmatis MC2 155 | MSMEG_1742 | - | 1e-129 | 63.56% (376) | oxidoreductase |
| M. thermoresistible (build 8) | TH_2872 | - | 7e-41 | 33.43% (359) | PUTATIVE 2Fe-2S iron-sulfur cluster binding domain protein |
| M. ulcerans Agy99 | MUL_4786 | - | 0.0 | 98.90% (365) | flavodoxin oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_1627 | - | 1e-117 | 61.25% (369) | oxidoreductase FAD-binding subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0335|M.marinum_M ---------MFTQT-------LTQTLAKRVLGSGLVDLLTGPHGIDRYTE
MUL_4786|M.ulcerans_Agy99 ---------MFTQT-------LTQTLAKRVLGSGLVDLLTGPHGIDRYTE
MAV_5168|M.avium_104 ---------MFTQTVQASGRVLARTVRQRVLGSELLDLLTGPHGVDRYTE
Mflv_4815|M.gilvum_PYR-GCK ---------MFTEV-----------LTKRSRRFALLDLLTGPHGVDRYTE
Mvan_1627|M.vanbaalenii_PYR-1 ---------MFTET-----------LTK-VRRSALLDLLTGPHGVDRYTE
MSMEG_1742|M.smegmatis_MC2_155 ---------MFTQTTAAG---FGRRLLAGSRMARLADLLTGPHGVDRYTE
MAB_1588c|M.abscessus_ATCC_199 -------MTFLTKATR-------RILTPDSSLGRLLTAALTPHAVDRYLE
Mb3259c|M.bovis_AF2122/97 MSKKHTTLNASIIDTRRPTVAGADRHPGWHALRKIAARITTPLLPDDYLH
Rv3230c|M.tuberculosis_H37Rv MSKKHTTLNASIIDTRRPTVAGADRHPGWHALRKIAARITTPLLPDDYLH
TH_2872|M.thermoresistible__bu MAKSEIKYSARVVDTVRPQVAGVENRPVLHRLRRFAERITTPLLPDDYLH
: * * * .
MMAR_0335|M.marinum_M LVEPTWTLGEARAKLIDVRRTTPRSVTLTLAPNDTFRSAMAAGTGPMAGQ
MUL_4786|M.ulcerans_Agy99 LVEPTWTLGEARAKLIDVRRTTPRSVTLTLAPNDTFRSAMAAGTGPLAGQ
MAV_5168|M.avium_104 LVAPTWTLGEARAKVTDVRRTTPRSVTLTLDPNDTFLSSHTV----TAGQ
Mflv_4815|M.gilvum_PYR-GCK LVDPTWTRGNARAKVVGVRRSTPRSVTLTLAPNRAFDG-------FRAGQ
Mvan_1627|M.vanbaalenii_PYR-1 LVDPTWTRGQARARVVGVRRSTPRSVTLTLAPNRAFAG-------FRAGQ
MSMEG_1742|M.smegmatis_MC2_155 LVEPTWSRHDARARVVAVRRQTPRSVTLTLEPNGAFTG-------FRAGQ
MAB_1588c|M.abscessus_ATCC_199 LVDPMITWEEARARVIRVQRRTTRSVTLTLRTTHQFKG-------FHAGQ
Mb3259c|M.bovis_AF2122/97 LANPLWSARELRGRILGVRRETEDSATLFIKPGWGFSFD------YQPGQ
Rv3230c|M.tuberculosis_H37Rv LANPLWSARELRGRILGVRRETEDSATLFIKPGWGFSFD------YQPGQ
TH_2872|M.thermoresistible__bu LINPLWSARELRGRVEDVRRETEDSATLVIKPGWGFSFD------YQPGQ
* * : : *.:: *:* * *.** : . * .**
MMAR_0335|M.marinum_M YVNLTVDIDGRRHTRCYSPAN----AEGS-ETLELTIGRHDGGLVSTYLC
MUL_4786|M.ulcerans_Agy99 YVNLTVDIDGRRHTRCYSPAN----AEGS-ETLELTIGRHDGGLVSTYLY
MAV_5168|M.avium_104 YVNLTVEIDGRRHTRCYSPAN----REGA-ATLELTIGRHDGGLVSNHLY
Mflv_4815|M.gilvum_PYR-GCK HINVSVEIDGRRRTRPYSPAN----AEGE-ANIELTIGRHDGGLVSTYLF
Mvan_1627|M.vanbaalenii_PYR-1 HVNVSVDIDGRRRTRPYSPAN----ADTD-PYLELTIGRHDGGLVSTYLF
MSMEG_1742|M.smegmatis_MC2_155 HINLSVEINGRRRTRCYSPAS----AEGA-EAIELTIGRHDGGLVSTYLC
MAB_1588c|M.abscessus_ATCC_199 FVQLGVVIDGVRHVRCFSPSC----ADDAREIIELTIARRPDGLVSNYLY
Mb3259c|M.bovis_AF2122/97 YIGIGLLVDGCWRWRSYSLTSSPAASG-SARMVTVTVKAMPEGFLSTHLV
Rv3230c|M.tuberculosis_H37Rv YIGIGLLVDGRWRWRSYSLTSSPAASG-SARMVTVTVKAMPEGFLSTHLV
TH_2872|M.thermoresistible__bu YIGIGLPVDGRWRWRSYSLTSPAVTSGRGGRTITITVKAMPEGFLSTHLV
.: : : ::* : * :* : : :*: *::*.:*
MMAR_0335|M.marinum_M DHARRGMVVGLAGVGGDFVLPQRRPRRILFVSGGSGITPVLAMLRTLVAQ
MUL_4786|M.ulcerans_Agy99 DHARRGMVVGLAGVGGDFVLPQRRPRRILFVSGGSGITPVLAMLRTLVAQ
MAV_5168|M.avium_104 DHARRGMVVGLAGVGGDFVLPDPRPRRVLFVSGGSGITPVMAMVRTLVSQ
Mflv_4815|M.gilvum_PYR-GCK DHARRGMVVGLDSVGGDFVLPARPAENILFVSGGSGITPVMSMLRTLRAR
Mvan_1627|M.vanbaalenii_PYR-1 ERARVGMVVGLDSVAGDFTLPAIPGP-TLFVSGGSGITPVMSMLRTLRAR
MSMEG_1742|M.smegmatis_MC2_155 DHAYAGMVLGLDSVGGDFVLPDTPPERILFVSGGSGITPVLSMLRTLRAQ
MAB_1588c|M.abscessus_ATCC_199 KHAAVGDVYSITPAAGTFVLPAPRPIRTLLIAAGSGITPVLSMARTLVGN
Mb3259c|M.bovis_AF2122/97 AGVKPGTIVRLAAPQGNFVLPDPAPPLILFLTAGSGITPVMSMLRTLVRR
Rv3230c|M.tuberculosis_H37Rv AGVKPGTIVRLAAPQGNFVLPDPAPPLILFLTAGSGITPVMSMLRTLVRR
TH_2872|M.thermoresistible__bu EGVKPGTIVRLAAPQGNFVMPNPAPPQVLFLTAGSGITPIMSMLRTLERR
. * : : * *.:* *:::.******:::* *** .
MMAR_0335|M.marinum_M HHLHIHGAEIAFIHYARNPAEACYRAELA----ALPGVRVLHGYTQSG-G
MUL_4786|M.ulcerans_Agy99 HHLHIHGAEIAFIHYAHNPAEACYRAELA----ALPGVRVLHGYTQSG-G
MAV_5168|M.avium_104 R----HRGEIAFIHYAPTPAEACYRDELA----ALPSVRVLHGYTRSA-G
Mflv_4815|M.gilvum_PYR-GCK R----HAGQVTFVHYARSADEACYRAELAVIAREMPNVTVLHGYTRVATD
Mvan_1627|M.vanbaalenii_PYR-1 R----HAGEVVFVHYARSAHEACYRDELADIASEMPNVTVLHGYTRDTTG
MSMEG_1742|M.smegmatis_MC2_155 A----FTGEVGFLHYARSADEACYRDELDGIRAAIPHLRVLHGYTRDAHG
MAB_1588c|M.abscessus_ATCC_199 G----YPGQLAVLYYAPTAADNAYAGELAALG-EVPSVTVRVEYTRDG--
Mb3259c|M.bovis_AF2122/97 N----QITDVVHLHSAPTAADVMFGAELAALAADHPGYRLSVRETRAQ--
Rv3230c|M.tuberculosis_H37Rv N----QITDVVHLHSAPTAADVMFGAELAALAADHPGYRLSVRETRAQ--
TH_2872|M.thermoresistible__bu D----QITDIVHIHSVPTPSDAMFSGELADLQRRHPSYRFRLRTTRTE--
:: :: . .. : : ** * . *:
MMAR_0335|M.marinum_M GDLVGRFGPDHLAAAMD--APDAVFVCGPTTLVDAVRENCEN------VF
MUL_4786|M.ulcerans_Agy99 GDLVGRFGPDHLAAAMD--APDAVFVCGPTTLVDAVRENCEN------VF
MAV_5168|M.avium_104 GDLSGRFGPDHLAAAMP--APDAVFVCGPTSLVDAVREHCDN------VY
Mflv_4815|M.gilvum_PYR-GCK SDLDGRFGAGQLPAGV--THADAVYACGPPALVDAVRELHPH------AE
Mvan_1627|M.vanbaalenii_PYR-1 SDLPPRFERAGIEP-------QAVYVCGPPALVDAVREVYPD------AA
MSMEG_1742|M.smegmatis_MC2_155 TDLVGRFGPDHLAAALPGNRPDAVYVCGPTDLVAAVEEHCDN------VR
MAB_1588c|M.abscessus_ATCC_199 ---GQHFSAEQLQAVAPWHADAQTFLCGPPSLHEAVSALYAERGLSDRLH
Mb3259c|M.bovis_AF2122/97 ----GRLDLTRIGQQVPDWRERQTWACGPEGVLNQADKVWSSAGASDRLH
Rv3230c|M.tuberculosis_H37Rv ----GRLDLTRIGQQVPDWRERQTWACGPEGVLNQADKVWSSAGASDRLH
TH_2872|M.thermoresistible__bu ----GRLDLSRLDDEVGDWRSRQTWACGPEGLLTAAERVWLGAGIGDRLH
:: : .: *** : .
MMAR_0335|M.marinum_M TESFVPAPIEAPAQPSGGRVRFADSG-IDVVDDGRSLLEQAESAGLAPEN
MUL_4786|M.ulcerans_Agy99 TESFVPAPIEAPAQPSGGRVRFADSG-IDVVDDGRSLLEQAESAGLAPEN
MAV_5168|M.avium_104 TESFTPPVLEASADPSGGRVTFSDSG-VDVVDDGRSLLEQAEAAGLSPQN
Mflv_4815|M.gilvum_PYR-GCK SESFVPPVFTAD-ESGGGTVSFTTAG-VDVVDDGRPLLNQAEDAGLAPES
Mvan_1627|M.vanbaalenii_PYR-1 SESFVPPVLSG--ESSGGRVTFTESG-VDVVDDGRPLLNQAEDAGLTPES
MSMEG_1742|M.smegmatis_MC2_155 SECFVPPTFSA--SATGGTVTFTDSA-VSVTDDGQPLLTQAEAAGLTPLS
MAB_1588c|M.abscessus_ATCC_199 TEEFTLATAGNT-GEAGGVLRFATSG-ITAENDGRPILEQAEAAGLQPEF
Mb3259c|M.bovis_AF2122/97 LERFAVSKTAPA--GAGGTVTFARSGKSVAADAATSLMDAGEGAGVQLPF
Rv3230c|M.tuberculosis_H37Rv LERFAVSKTAPA--GAGGTVTFARSGKSVAADAATSLMDAGEGAGVQLPF
TH_2872|M.thermoresistible__bu LERFAVARVANP--GGGGTIEFARSGKSTVVDAATSLMDAGEQAGVQMPF
* *. . ** : *: :. . : . .:: .* **:
MMAR_0335|M.marinum_M GCRMGICHTCTRRKTSGTVRNLVTGAVSVAPDEDVQVCVSVPVGDVDLSL
MUL_4786|M.ulcerans_Agy99 GCRMGICHTCTRRKTSGTVRNLVTGAVSVAPDEDVQICVSVPVGDVDLSL
MAV_5168|M.avium_104 GCRMGICHTCTRRKTAGTVRNLVTGAVSTAPDEDVQICVSVPVGDVDLSL
Mflv_4815|M.gilvum_PYR-GCK GCRMGICFSCTRRKTSGVVRNVVTGAVSSTEEEDVQICVSAAVGDVDIDL
Mvan_1627|M.vanbaalenii_PYR-1 GCRMGICFSCTRRKTSGAVRNVITGVVSSTEEEDVQICVSAPVGDVDVAL
MSMEG_1742|M.smegmatis_MC2_155 GCRMGICHSCTRTKTRGAVRNLITGAVSGTDNEDIQICVTAPVGDVDIDL
MAB_1588c|M.abscessus_ATCC_199 GCRMGICFACTAVKTSGCTRNLRTGDENDDPDQHIQLCITAPVGDVSINL
Mb3259c|M.bovis_AF2122/97 GCRMGICQSCVVDLVEGHVRDLRTG-QRHEPGTRVQTCVSAASGDCVLDI
Rv3230c|M.tuberculosis_H37Rv GCRMGICQSCVVDLVEGHVRDLRTG-QRHEPGTRVQTCVSAASGDCVLDI
TH_2872|M.thermoresistible__bu GCRMGICQSCVIGLVDGHVRDLRTG-EVHDPGTRVQTCVSAAAGDCVLDI
******* :*. . * .*:: ** :* *::.. ** : :