For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MMAVPSSRTPQVAMVGGGQLARMTHQAAIALGQNLRVLVNSADDPAAQVTPNVVIGSHTDLDDLRRIAAG ADVVTFDHEHVPAELLDKLVAEGVVLAPPPQALVHAQDKLVMRRRLQALDVPVPRYLGIHQLEELGELDA FARELDAPLVVKAVRGGYDGRGVQMARDLADARDIASGYLAGGVPVLVEERVQLRRELSALVARSPFGQG AAWPVVETVQQDGICVAVIAPAPALSAQAGADAQQLALRLAAELGVVGVLAVELFETTDGALVVNELAMR PHNSGHWTMDGSRTSQFEQHLRAVLDYPLGDTEAIAPVTVMTNVLGAAAQPAMSLDERLHHLFARMPDAR VHLYGKDERPGRKVGHINFLGFDPAAVAGLRERAELAAHWLSHGQWTDGWDPHG
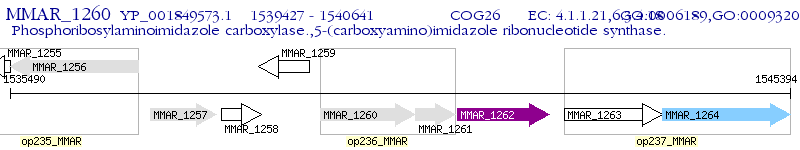
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1260 | purK | - | 100% (404) | phosphoribosylaminoimidazole carboxylase ATPase subunit PurK (air carboxylase) (AirC) |
| M. marinum M | MMAR_0686 | purT | 4e-14 | 26.50% (400) | formate-dependent phosphoribosylglycinamide |
| M. marinum M | MMAR_3848 | accA1 | 5e-05 | 28.96% (183) | acetyl-/propionyl-coenzyme A carboxylase alpha chain AccA1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3304c | purK | 0.0 | 80.05% (411) | phosphoribosylaminoimidazole carboxylase ATPase subunit |
| M. gilvum PYR-GCK | Mflv_4747 | - | 1e-162 | 70.53% (397) | phosphoribosylaminoimidazole carboxylase ATPase subunit |
| M. tuberculosis H37Rv | Rv3276c | purK | 0.0 | 80.05% (411) | phosphoribosylaminoimidazole carboxylase ATPase subunit |
| M. leprae Br4923 | MLBr_00735 | purK | 1e-180 | 76.92% (416) | phosphoribosylaminoimidazole carboxylase ATPase subunit |
| M. abscessus ATCC 19977 | MAB_3620c | - | 1e-153 | 66.00% (403) | phosphoribosylaminoimidazole carboxylase ATPase subunit |
| M. avium 104 | MAV_4245 | purK | 0.0 | 81.22% (394) | phosphoribosylaminoimidazole carboxylase ATPase subunit |
| M. smegmatis MC2 155 | MSMEG_1819 | purK | 1e-159 | 68.42% (399) | phosphoribosylaminoimidazole carboxylase ATPase subunit |
| M. thermoresistible (build 8) | TH_0889 | purK | 1e-169 | 75.00% (392) | PROBABLE PHOSPHORIBOSYLAMINOIMIDAZOLE CARBOXYLASE ATPASE |
| M. ulcerans Agy99 | MUL_2628 | purK | 0.0 | 99.75% (404) | phosphoribosylaminoimidazole carboxylase ATPase subunit |
| M. vanbaalenii PYR-1 | Mvan_1713 | - | 1e-161 | 71.07% (394) | phosphoribosylaminoimidazole carboxylase ATPase subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4747|M.gilvum_PYR-GCK --MPHPAVTAGQRCGVGHDTMYAVSRRPSSP------PVVAMIGGGQLAR
Mvan_1713|M.vanbaalenii_PYR-1 --------------------MYAVSRTPASP------PVVAMIGGGQLAR
MMAR_1260|M.marinum_M --------------------MMAVPSSRT--------PQVAMVGGGQLAR
MUL_2628|M.ulcerans_Agy99 MIHNDLVLQPPNTPVMGHDTMMAVPSSRT--------PQVAMVGGGQLAR
Mb3304c|M.bovis_AF2122/97 --------------------MMAVASSRTPAVTSFIAPLVAMVGGGQLAR
Rv3276c|M.tuberculosis_H37Rv --------------------MMAVASSRTPAVTSFIAPLVAMVGGGQLAR
MAV_4245|M.avium_104 -----------------------------------------MVGGGQLAR
MLBr_00735|M.leprae_Br4923 --------------------MMAVPSRCSLG----VAPLVAMVGGGQLAR
TH_0889|M.thermoresistible__bu ---------------------------------------VAMVGGGQLAR
MSMEG_1819|M.smegmatis_MC2_155 --------------------MPRASQTNSSA-----VPVVAMIGGGQLAR
MAB_3620c|M.abscessus_ATCC_199 --------------------MSAT-------------PVVTMIGGGQLAR
*:*******
Mflv_4747|M.gilvum_PYR-GCK MTAQAAIALGQTLRVLAVRSDESAAQVTPDVVLGSHTDLDALRRVADGAT
Mvan_1713|M.vanbaalenii_PYR-1 MTAQAAIALGQTLRVLAVRSDESAAQVTPDVVLGSHTDLDALRRVADGAT
MMAR_1260|M.marinum_M MTHQAAIALGQNLRVLVNSADDPAAQVTPNVVIGSHTDLDDLRRIAAGAD
MUL_2628|M.ulcerans_Agy99 MTHQAAIALGQNLRVLVNSADDPAAQVTPNVVIGSHTDLDDLRRIAAGAD
Mb3304c|M.bovis_AF2122/97 MTHQAAIALGQNLRVLVTSADDPAAQVTPNVVIGSHTDLAALRRVAAGAD
Rv3276c|M.tuberculosis_H37Rv MTHQAAIALGQNLRVLVTSADDPAAQVTPNVVIGSHTDLAALRRVAAGAD
MAV_4245|M.avium_104 MTHQAAIALGQSLRVLATAADESAALVASDVVIGSHTDLEDLRRVAAGAD
MLBr_00735|M.leprae_Br4923 MTHQAAIALGQTLRVLATAADEPAAQVTPDVVIGSHTDLEDLRRVALGAD
TH_0889|M.thermoresistible__bu MTHQAAIALGQTLRVLAVDSDESAAQVTPDVVLGSHTDIDALRRAAAGAQ
MSMEG_1819|M.smegmatis_MC2_155 MTHQAAIALGQTLRVLAVDPNDPAAQVSPDIVLGSHTDYDALVKVAQGAT
MAB_3620c|M.abscessus_ATCC_199 MTHQASIALGQTLRVLAHAADEPAAQVTPDIVLGSHTELDDLRRAAKDAT
** **:*****.****. .::.** *:.::*:****: * : * .*
Mflv_4747|M.gilvum_PYR-GCK AFTFDHEHVPTEHLDVLIAEGVNVAPPPEALIHAQDKLLMRRRLSELGAP
Mvan_1713|M.vanbaalenii_PYR-1 AFTFDHEHVPPALLEQLVADGVNVLPPPSALVHAQDKLLMRERLSSLGVP
MMAR_1260|M.marinum_M VVTFDHEHVPAELLDKLVAEGVVLAPPPQALVHAQDKLVMRRRLQALDVP
MUL_2628|M.ulcerans_Agy99 VVTFDHEHVPAELLDKLVAEGVVLAPPPQALVHAQDKLVMRRRLQALDVP
Mb3304c|M.bovis_AF2122/97 VLTFDHEHVPNELLEKLVADGVNVAPSPQALVHAQDKLVMRQRLAAAGVA
Rv3276c|M.tuberculosis_H37Rv VLTFDHEHVPNELLEKLVADGVNVAPSPQALVHAQDKLVMRQRLAAAGVA
MAV_4245|M.avium_104 VLTFDHEHVPGELLDKLVAEGVNVAPPPQALVHAQDKLVMRRRLESLGVP
MLBr_00735|M.leprae_Br4923 ALTFDHEHVPTELLDKLVAEGINVAPSPQALVHAQDKLVMRRRLAALGAA
TH_0889|M.thermoresistible__bu VLTFDHEHVPTELLEKLQAEGVNVAPPPQALIHAQDKLVMRRRLEALGAP
MSMEG_1819|M.smegmatis_MC2_155 VVTFDHEHVPTEHLDRLVADGVTVAPPPEALVHAQDKLVMRRRLADLGAP
MAB_3620c|M.abscessus_ATCC_199 VVTFDHEHVPTAFLETLQGEGVNVQPPPSALIYAQDKTLMRRKLRDIGAP
..******** *: * .:*: : *.*.**::**** :**.:* ...
Mflv_4747|M.gilvum_PYR-GCK VPRFSAVS---SVEDAVAFSEQV----GGPAVIKTVRGGYDGRGVVIASD
Mvan_1713|M.vanbaalenii_PYR-1 VPRYTAVG---SVDDAVAFCTEL----GGPVVIKTVRGGYDGRGVIMASS
MMAR_1260|M.marinum_M VPRYLGIHQLEELGELDAFAREL----DAPLVVKAVRGGYDGRGVQMARD
MUL_2628|M.ulcerans_Agy99 VPRYLGIHQLEELGELDAFARQL----DAPLVVKAVRGGYDGRGVQMARD
Mb3304c|M.bovis_AF2122/97 VPRYAGIK---DPDEIDVFAARV----DAPIVVKAVRGGYDGRGVRMARD
Rv3276c|M.tuberculosis_H37Rv VPRYAGIK---DPDEIDVFAARV----DAPIVVKAVRGGYDGRGVRMARD
MAV_4245|M.avium_104 VPRYAEIN---SLDELDAFARRV----GGPVVVKAVRGGYDGRGVRMARD
MLBr_00735|M.leprae_Br4923 MPRFMALDSVDDLAEIDAFAQRLTGSKDAPMVVKAVRGGYDGRGVQMVRD
TH_0889|M.thermoresistible__bu VPRFAAVD---DVDDLDDFARRV----DAPLVLKTIRGGYDGRGVILTTD
MSMEG_1819|M.smegmatis_MC2_155 IPQFAEVT---AADEAQEFSREV----GGAAVIKTIRGGYDGRGVVLLDT
MAB_3620c|M.abscessus_ATCC_199 VPGFADIR---AVSDVERFAAET----GSHVVLKAVRGGYDGRGVWITDT
:* : : : *. . .. *:*::********* :
Mflv_4747|M.gilvum_PYR-GCK TDEVRSAVEGYLADGVPVMIEERVAMRRELAALVARSPFGQGAAWPVVET
Mvan_1713|M.vanbaalenii_PYR-1 VAEVRSVVSQYLDDGVPVLIEERVAMRRELAALVARSPFGQGAAWPVVET
MMAR_1260|M.marinum_M LADARDIASGYLAGGVPVLVEERVQLRRELSALVARSPFGQGAAWPVVET
MUL_2628|M.ulcerans_Agy99 LADARDIASGYLAGGVPVLVEERVQLRRELSALVARSPFGQGAAWPVVET
Mb3304c|M.bovis_AF2122/97 VADARDFARECLADGVAVLVEERVDLRRELSALVARSPFGQGAAWPVVQT
Rv3276c|M.tuberculosis_H37Rv VADARDFARECLADGVAVLVEERVDLRRELSALVARSPFGQGAAWPVVQT
MAV_4245|M.avium_104 PVHAREIAAAFLADGVPVLAEEQVSLRRELSALVARSPFGQGAAWPVVET
MLBr_00735|M.leprae_Br4923 SAHAREVASGYLVDGMPVLVEERVELRRELSALVARSPFGQGAAWPVVET
TH_0889|M.thermoresistible__bu LPEAREAARRYLADGVPVLVEERVAMRRELTALVARSPFGQGAAWPVVET
MSMEG_1819|M.smegmatis_MC2_155 PEQAREVAAKYLTDGVPILIEERVAMRRELSALVARSAFGQGSAWPVVQT
MAB_3620c|M.abscessus_ATCC_199 LDEARTVAAEQLAAGVELLAEERVDMRRELSAMVARSPFGQGAAWPVVET
..* . * *: :: **:* :****:*:****.****:*****:*
Mflv_4747|M.gilvum_PYR-GCK VQRDGICVTVLAPAPDISDELGSAASELALHIASSLGVVGVLAVELFETV
Mvan_1713|M.vanbaalenii_PYR-1 VQRDGICVTVLAPAPGLSDELGSAASELALSVASSLGVVGVLAVELFETV
MMAR_1260|M.marinum_M VQQDGICVAVIAPAPALSAQAGADAQQLALRLAAELGVVGVLAVELFETT
MUL_2628|M.ulcerans_Agy99 VQQDGICVAVIAPAPALSAQAGADAQQLALRLAAELGVVGVLAVELFETT
Mb3304c|M.bovis_AF2122/97 VQRDGTCVLVIAPAPALPDDLATAAQRLALQLADELGVVGVLAVELFETT
Rv3276c|M.tuberculosis_H37Rv VQRDGTCVLVIAPAPALPDDLATAAQRLALQLADELGVVGVLAVELFETT
MAV_4245|M.avium_104 VQRDGICVQVIAPAPGLGEDLAADAQRLALRLAGELGVVGVLAVELFETT
MLBr_00735|M.leprae_Br4923 VQRDGICVLVVAPALALADDLASAAQQLALRLAAELGVVGVFAVELFETA
TH_0889|M.thermoresistible__bu VQRDGICVEVLAPAPGLPDEVGAAAQELALRLAGELGVVGLLAVELFETA
MSMEG_1819|M.smegmatis_MC2_155 VQRDGICVEVIAPAPDLPDDLAASAQQLGLRLAAELGVIGVLAVELFETV
MAB_3620c|M.abscessus_ATCC_199 VQREGICAVVIAPAPQLPEELALQAEQLALRIANELGVTGCLAVELFETN
**::* *. *:*** : : . *..*.* :* .*** * :*******
Mflv_4747|M.gilvum_PYR-GCK DGGLLVNELAMRPHNSGHWTMDGARTSQFEQHLRAVLDYPLGDTSALAP-
Mvan_1713|M.vanbaalenii_PYR-1 SGGLLVNELAMRPHNSGHWTMDGARTSQFEQHLRAVLDYPLGDTSALAP-
MMAR_1260|M.marinum_M DGALVVNELAMRPHNSGHWTMDGSRTSQFEQHLRAVLDYPLGDTEAIAPV
MUL_2628|M.ulcerans_Agy99 DGALVVNELAMRPHNSGHWTMDGSRTSQFEQHLRAVLDYPLGDTEAIAPV
Mb3304c|M.bovis_AF2122/97 DGALLVNELAMRPHNSGHWTIDGARTSQFEQHLRAVLDYPLGDSDAVVPV
Rv3276c|M.tuberculosis_H37Rv DGALLVNELAMRPHNSGHWTIDGARTSQFEQHLRAVLDYPLGDSDAVVPV
MAV_4245|M.avium_104 DGALLVNELAMRPHNSGHWTMDGSRTGQFEQHLRAVLDYPLGDTDALAPV
MLBr_00735|M.leprae_Br4923 DGALLVNELAMRPHNSGHWTMDGARTSQFEQHLRAVLDYPLGETDAVAPV
TH_0889|M.thermoresistible__bu EGGLMVNELAMRPHNSGHWTMNGAVTSQFEQHLRAVLDYPLGDTSAIAPV
MSMEG_1819|M.smegmatis_MC2_155 DGRILVNELAMRPHNSGHWTMDGAVTSQFEQHLRAVLDYPLGETSARAEI
MAB_3620c|M.abscessus_ATCC_199 DGRLLVNELAMRPHNSGHWSMDGARTGQFEQHLRAVLDYPLGSTEPLAPV
.* ::**************:::*: *.***************.:.. .
Mflv_4747|M.gilvum_PYR-GCK TVMGNVLGAPETPAMS---MDERVHHLFARMPDAKVHLYGKEERPGRKIG
Mvan_1713|M.vanbaalenii_PYR-1 TAMGNVLGAPVTPAMS---MDERVHHLFARMPDAKVHLYGKEERPGRKIG
MMAR_1260|M.marinum_M TVMTNVLGAAAQPAMS---LDERLHHLFARMPDARVHLYGKDERPGRKVG
MUL_2628|M.ulcerans_Agy99 TVMTNVLGAAAQPAMS---LDERLHHLFARMPDARVHLYGKDERPGRKVG
Mb3304c|M.bovis_AF2122/97 TVMANVLGAAQPPAMS---VDERLHHLFARMPDARVHLYGKAERPGRKVG
Rv3276c|M.tuberculosis_H37Rv TVMANVLGAAQPPAMS---VDERLHHLFARMPDARVHLYGKAERPGRKVG
MAV_4245|M.avium_104 TVMANVLGARERPRMS---VDERLHHLFARMPDARVHLYGKGERPGRKVG
MLBr_00735|M.leprae_Br4923 TVMVNVLGAPQPPTLSVVTMDERLHHLFARMPDARVHLYDKVERPGRKVG
TH_0889|M.thermoresistible__bu TVMANVLGAPQTPAMS---MDERLHHLFGRIPDARVHLYGKQERPGRKLG
MSMEG_1819|M.smegmatis_MC2_155 VVMSNVLGAPETPAMS---LDERIHHLFARMPDAKVHMYGKGERPGRKLG
MAB_3620c|M.abscessus_ATCC_199 TVMANVLGAPQTPAMS---LDERMHHLMGRIPEAKVHLYGKGERPGRKLG
..* ***** * :* :***:***:.*:*:*:**:*.* ******:*
Mflv_4747|M.gilvum_PYR-GCK HVNIVGGP------SEPVESLRERAERAAHWLSHAQWTDGWDSHA-----
Mvan_1713|M.vanbaalenii_PYR-1 HVNIVGG-------TESVESLRERAERAAHWLSHAEWTDGWDPHA-----
MMAR_1260|M.marinum_M HINFLG------FDPAAVAGLRERAELAAHWLSHGQWTDGWDPHG-----
MUL_2628|M.ulcerans_Agy99 HINFLG------FDPAAVAGLRERAELAAHWLSHGQWTDGWDPHG-----
Mb3304c|M.bovis_AF2122/97 HINFLG------SD---VAQLCERAELAAHWLSHGRWTDGWDPHRASDDA
Rv3276c|M.tuberculosis_H37Rv HINFLG------SD---VAQLCERAELAAHWLSHGRWTDGWDPHRASDDA
MAV_4245|M.avium_104 HINFLGDFPA--ADTAGLAALRRRAELAAHWLSHGQWTDGWDPHEQERHQ
MLBr_00735|M.leprae_Br4923 HINFRG--TD--KDRKNPTKLRERAELAAHWLSHGQWTDGWDPHRAGDDV
TH_0889|M.thermoresistible__bu HVNVVG---------SDLAVVRERAGRAAHWLAHAQWTDGWDGHAD----
MSMEG_1819|M.smegmatis_MC2_155 HVNLVG------KPGEDVAAVRERANRASHWLSHAQWTDNWNEHGTDTE-
MAB_3620c|M.abscessus_ATCC_199 HVNIQGREDGSLADPGYVAEVRERAERAAHWLSHAVWTDGWEGH------
*:*. * : .** *:***:*. ***.*: *
Mflv_4747|M.gilvum_PYR-GCK ------------------
Mvan_1713|M.vanbaalenii_PYR-1 ------------------
MMAR_1260|M.marinum_M ------------------
MUL_2628|M.ulcerans_Agy99 ------------------
Mb3304c|M.bovis_AF2122/97 VGVPPACGGRSDEEERRL
Rv3276c|M.tuberculosis_H37Rv VGVPPACGGRSDEEERRL
MAV_4245|M.avium_104 ------------------
MLBr_00735|M.leprae_Br4923 VEISLACGGRNDAQRQR-
TH_0889|M.thermoresistible__bu ------------------
MSMEG_1819|M.smegmatis_MC2_155 ------------------
MAB_3620c|M.abscessus_ATCC_199 ------------------