For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAAQTGSLDGRVAFVTGAARAQGRSHAVRLAREGADIIAMDICGPVSETITYAPATAADLSETIRAVESE GRKVLARQADVRDSAALQQLVADGVEEFGRLDVVVANAGVLGWGRLWELTDEQWDTVIGVNLSGTWRTLR AAVPAMIEAGNGGSIIVVSSSAGLKATPGNGHYAASKHGLVALTNTLAIELGEYDIRVNSIHPYSVETPM IEPDVMMQVFGEHPRFLHSFPPMPLQYKGLMTSEEVSDVVVWLAGDGSGTLSGAQIPVDKGALKY
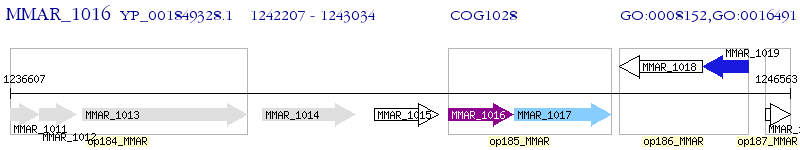
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1016 | - | - | 100% (275) | short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4754 | - | 2e-64 | 46.01% (276) | short-chain type dehydrogenase/reductase |
| M. marinum M | MMAR_1965 | fabG | 2e-56 | 42.39% (276) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0706 | fabG | 1e-132 | 81.82% (275) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_5072 | fabG | 1e-113 | 69.31% (277) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. tuberculosis H37Rv | Rv0687 | fabG | 1e-132 | 81.82% (275) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01740 | - | 3e-30 | 35.32% (218) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3844c | - | 3e-99 | 62.31% (268) | putative short-chain dehydrogenase/reductase |
| M. avium 104 | MAV_4486 | - | 1e-129 | 80.00% (275) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1410 | fabG | 1e-118 | 73.65% (277) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. thermoresistible (build 8) | TH_3434 | - | 1e-118 | 74.73% (273) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0768 | - | 1e-138 | 98.39% (248) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_1286 | fabG | 1e-116 | 69.86% (282) | 3-ketoacyl-(acyl-carrier-protein) reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5072|M.gilvum_PYR-GCK MPMSSGTPLEGRVALITGAARGQGRAHAIRLAADGADIVALDVCKPVSES
Mvan_1286|M.vanbaalenii_PYR-1 MPMASAAPLEGRVALITGAARGQGRAHAVRLAAEGADIIAIDVCGPVSES
MSMEG_1410|M.smegmatis_MC2_155 -MATVGKTLEGRVAFVTGAARGQGRAHAVRLADEGADIIAIDVCAPVDDT
TH_3434|M.thermoresistible__bu ------------VAFITGAARGQGRAHAIRLAEEGADIIAIDVCGPVSDT
MMAR_1016|M.marinum_M -MAAQTGSLDGRVAFVTGAARAQGRSHAVRLAREGADIIAMDICGPVSET
MUL_0768|M.ulcerans_Agy99 ----------------------------MRLAREGADIIAMDICGPVSES
Mb0706|M.bovis_AF2122/97 -MSARGGSLHGRVAFVTGAARAQGRSHAVRLAREGADIVALDICAPVSGS
Rv0687|M.tuberculosis_H37Rv -MSARGGSLHGRVAFVTGAARAQGRSHAVRLAREGADIVALDICAPVSGS
MAV_4486|M.avium_104 -MSGQAGSLQGRVAFITGAARGQGRSHAVRLAAEGADIIACDICAPVSAS
MAB_3844c|M.abscessus_ATCC_199 ---MADQPLAGKVAFVTGAARGQGRQHAVRLAQAGADVVAIDACAPVSEY
MLBr_01740|M.leprae_Br4923 -MVKTAQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDR-------
:*** **:: *
Mflv_5072|M.gilvum_PYR-GCK VTYPMPTSEDLAETVRLVEETGRKVLAR-EVDIRDLAAQQQLVADAVEQF
Mvan_1286|M.vanbaalenii_PYR-1 ITYSMPTSEDLAETVRLVEATGRKVLAR-EVDIRDLAAQQQVVADGVEQF
MSMEG_1410|M.smegmatis_MC2_155 ITYPSATPEDLAETVAAVEATGRKVLAR-EVDIRDLAAQQQVVADGVEQF
TH_3434|M.thermoresistible__bu ITYPPATEDDLAETVRAVEATGRKVLAR-TADIRDLAAQQQIVAEGMERF
MMAR_1016|M.marinum_M ITYAPATAADLSETIRAVESEGRKVLAR-QADVRDSAALQQLVADGVEEF
MUL_0768|M.ulcerans_Agy99 ITYAPATAADLSETIRAVESEGRKVLAR-QADVRDSAALQQLVADGVEEF
Mb0706|M.bovis_AF2122/97 VTYPPATSEDLGETVRAVEAEGRKVLAR-EVDIRDDAELRRLVADGVEQF
Rv0687|M.tuberculosis_H37Rv VTYPPATSEDLGETVRAVEAEGRKVLAR-EVDIRDDAELRRLVADGVEQF
MAV_4486|M.avium_104 VTYAPASPEDLDETARLVEDQGRKALTR-VLDVRDDAALRELVADGMEQF
MAB_3844c|M.abscessus_ATCC_199 AGYEAATPEDLKETVRLVEAAGRKIIAE-QADVRNGAALQSVVEEAVAQF
MLBr_01740|M.leprae_Br4923 ------HCDGLAQTVSEARALGAQVPEYRALDISDYDEVAAFGADIHARH
.* :* .. * : *: : . : ..
Mflv_5072|M.gilvum_PYR-GCK GRLDIVVANAGVLSWGRIWEMSEDQWDTVVDVNLNGTWRTIRAAVPAMIE
Mvan_1286|M.vanbaalenii_PYR-1 GRLDIVVANAGVLSWGRIWEMSEEQWDTVVDVNLNGTWRTVRAAVPAMIE
MSMEG_1410|M.smegmatis_MC2_155 GRLDIVVANAGVLSWGRIFEMSPEQWDTVVDVNLNGTWRTIRAAVPAMIE
TH_3434|M.thermoresistible__bu GRLDIVVANAGVLSWGRLWEMSEEQWDTVIDVNLNGTWRTIRAVVPAMIE
MMAR_1016|M.marinum_M GRLDVVVANAGVLGWGRLWELTDEQWDTVIGVNLSGTWRTLRAAVPAMIE
MUL_0768|M.ulcerans_Agy99 GRLDVMVANAGVFGWGRLWELTDEQWDTVIGVNLSGTWRTLRAAVPAMIE
Mb0706|M.bovis_AF2122/97 GRLDIVVANAGVLGWGRLWELTDEQWETVIGVNLTGTWRTLRATVPAMID
Rv0687|M.tuberculosis_H37Rv GRLDIVVANAGVLGWGRLWELTDEQWETVIGVNLTGTWRTLRATVPAMID
MAV_4486|M.avium_104 GRLDVVVANAGVLSWGRVWELTDEQWDTVIGVNLTGTWRTLRATVPAMIE
MAB_3844c|M.abscessus_ATCC_199 GHIDILIANAGVLSWGRLWEMSDQQWEDIIDTNLTGTWKTVKAVVPTMIE
MLBr_01740|M.leprae_Br4923 PSMDIVMNIAGVSVWGTVDRLTHDQWSNVVAINLMGPIHIIETFVPAILA
:*::: *** ** : .:: :**. :: ** *. : :.: **:::
Mflv_5072|M.gilvum_PYR-GCK AGNGGSIIIVSSSAGLKATPGNAHYAASKHGLVALTNALALEAGEFGIRV
Mvan_1286|M.vanbaalenii_PYR-1 AGNGGSIVIVSSSAGLKATPGNAHYAASKHGLVALTNALAIEAGEFGIRV
MSMEG_1410|M.smegmatis_MC2_155 AGNGGSIIIVSSSAGLKATPGNGHYAASKHGLVALTNSLALEVGEYGIRV
TH_3434|M.thermoresistible__bu AGNGGSIIIVSSSAGLKATPGNGHYAASKHGLVALTNALAIEVGEYGIRV
MMAR_1016|M.marinum_M AGNGGSIIVVSSSAGLKATPGNGHYAASKHGLVALTNTLAIELGEYDIRV
MUL_0768|M.ulcerans_Agy99 AGNGGSIIVVSSSAGLKATPGNGHYAASKHGLVALTNTLAIELGEYDIGV
Mb0706|M.bovis_AF2122/97 AGNGGSIVVVSSSAGLKATPGNGHYAASKHALVALTNTLAIELGEFGIRV
Rv0687|M.tuberculosis_H37Rv AGNGGSIVVVSSSAGLKATPGNGHYAASKHALVALTNTLAIELGEFGIRV
MAV_4486|M.avium_104 AGNGGSIVVVSSSAGLKATPGNGHYSASKHGLTALTNTLAIELGEYGIRV
MAB_3844c|M.abscessus_ATCC_199 AGNGGSIVIVSSVSGLKGTPGNGAYVASKFGLTGLTKSLTIELAEYGIRV
MLBr_01740|M.leprae_Br4923 AGRGGHLVNVSSAAGLVALPWHAAYSASKYGLRGLSEVLRFDLARYRIGV
**.** :: *** :** . * :. * ***..* .*:: * :: ..: * *
Mflv_5072|M.gilvum_PYR-GCK NSIHPYSIDTPMIEKDAMMEIFSKYPTFLHSFAPMPFKPVARDGKPGLQE
Mvan_1286|M.vanbaalenii_PYR-1 NSIHPYSIDTPMIEKDAMMEIFSKYPTYLHSFAPMPFKPVAREGKPGLQE
MSMEG_1410|M.smegmatis_MC2_155 NSIHPYSIDTPMIEPEAMAQVFAKYPNYLHSFAPMPYQPVG-EGAEGLST
TH_3434|M.thermoresistible__bu NSIHPYSIDTPMIERDAMMAVFAKHPNYLHSFAPMPLHPVNREGDGGRQE
MMAR_1016|M.marinum_M NSIHPYSVETPMIEPDVMMQVFGEHPRFLHSFPPMPLQYKG---------
MUL_0768|M.ulcerans_Agy99 NSIHPYSVETPMIEPDVMMQVFGEHPRFLHSFPPMPLQYKG---------
Mb0706|M.bovis_AF2122/97 NSIHPYSVDTPMIEPEAMIQTFAKHPGYVHSFPPMPLQPKG---------
Rv0687|M.tuberculosis_H37Rv NSIHPYSVDTPMIEPEAMIQTFAKHPGYVHSFPPMPLQPKG---------
MAV_4486|M.avium_104 NSIHPYSVETPMIEPEAMMEIFARHPSFVHSFPPMPVQPNG---------
MAB_3844c|M.abscessus_ATCC_199 NSIHPYGVTTPMITGDAMMKILAEHPNYLPSIAPMPLSPTK---------
MLBr_01740|M.leprae_Br4923 SVVVPGAVRTPLVDTIEIAGVDRQDPRFSRWVDRFRGHAVSAERAAEKIL
. : * .: **:: : . * : . :
Mflv_5072|M.gilvum_PYR-GCK FMTVE-----------------------------EVSDVVAWLAGDSSAT
Mvan_1286|M.vanbaalenii_PYR-1 FMTVE-----------------------------EVADVVAWLAGDASST
MSMEG_1410|M.smegmatis_MC2_155 FMTPE-----------------------------EVSDVVAWLAGDGSAT
TH_3434|M.thermoresistible__bu FMTPE-----------------------------EVADVVAWLAGDGSAT
MMAR_1016|M.marinum_M LMTSE-----------------------------EVSDVVVWLAGDGSGT
MUL_0768|M.ulcerans_Agy99 LMTSE-----------------------------EVSDVVVWLAGDGSGT
Mb0706|M.bovis_AF2122/97 FMTPD-----------------------------EISDVVVWLAGDGSGA
Rv0687|M.tuberculosis_H37Rv FMTPD-----------------------------EISDVVVWLAGDGSGA
MAV_4486|M.avium_104 FMTAD-----------------------------EVADVVAWLAGDGSGT
MAB_3844c|M.abscessus_ATCC_199 MLEPD-----------------------------EISDVVLWLASDASGT
MLBr_01740|M.leprae_Br4923 AGVAKNRYLIYTSHDIRVLYGFKRLAWWPYGVVMKQVNVVFTRALRPSPA
. : :** * * :
Mflv_5072|M.gilvum_PYR-GCK ISGSQIAVDRGTMKY-----------
Mvan_1286|M.vanbaalenii_PYR-1 ISGSQIAVDRGTMKY-----------
MSMEG_1410|M.smegmatis_MC2_155 LSGSQIAVDRGVMKY-----------
TH_3434|M.thermoresistible__bu LSGSQIAVDRGVMKY-----------
MMAR_1016|M.marinum_M LSGAQIPVDKGALKY-----------
MUL_0768|M.ulcerans_Agy99 LSGAQIPVDKGALKY-----------
Mb0706|M.bovis_AF2122/97 LSGNQIPVDKGALKY-----------
Rv0687|M.tuberculosis_H37Rv LSGNQIPVDKGALKY-----------
MAV_4486|M.avium_104 LTGTQIPVDKGALKY-----------
MAB_3844c|M.abscessus_ATCC_199 LAGAQVPVDKGHLTY-----------
MLBr_01740|M.leprae_Br4923 TMRCVEPIKVGVYSQPRSGEGGKSGN
.:. * .