For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTTKHREVAKLDRVPLPVEAARVAVTGWQVTRTAARFVTKLPGKGFASSWQQKIIKEIPQTFADLGPTYV KFGQIIASSPGAFGESLSREFRGLLDRVPPADTDQVHKLFVEELGAEPAELFAAFEEEPFASASIAQVHY ATLKTGEEVVVKIQRPGIRRRVAADLQILQRFAQAVELAKLGRRLSAQDVVADFSDNLAEELDFRLEAQS MDAWVAHLHASPLGRNIRVPQVHWNFTSERVLTMERVHGVRIDNVAAIRKAGFDGTELVKALLFSVFEGG LRHGLFHGDLHAGNLYVDEEGRIVFFDFGIMGRIDPRTRWLLRELVYALLVKKDHAAAGKIVVLMGAVGT MKPEAQAAKDLEKFATPLTMQSLGDMSYADIGRQLSALADAYDVKLPRELVLIGKQFLYVERYMKLLAPK WQMMSDPQLTGYFANFMVEVSREHQGDSSDASGDASDVEA*
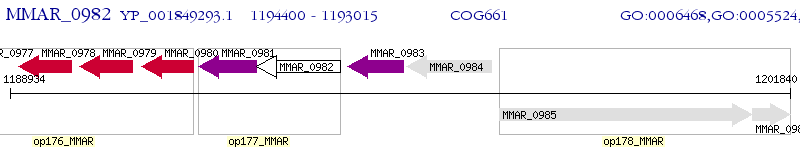
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0982 | - | - | 100% (461) | hypothetical protein MMAR_0982 |
| M. marinum M | MMAR_1366 | - | 1e-24 | 31.12% (286) | ABC transporter ATP-binding protein |
| M. marinum M | MMAR_1443 | - | 1e-22 | 28.39% (317) | hypothetical protein MMAR_1443 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0666c | - | 0.0 | 91.52% (448) | hypothetical protein Mb0666c |
| M. gilvum PYR-GCK | Mflv_5111 | - | 0.0 | 82.63% (449) | hypothetical protein Mflv_5111 |
| M. tuberculosis H37Rv | Rv0647c | - | 0.0 | 91.52% (448) | hypothetical protein Rv0647c |
| M. leprae Br4923 | MLBr_01898 | - | 0.0 | 83.48% (448) | hypothetical protein MLBr_01898 |
| M. abscessus ATCC 19977 | MAB_3879 | - | 0.0 | 72.32% (448) | hypothetical protein MAB_3879 |
| M. avium 104 | MAV_4514 | - | 0.0 | 90.23% (440) | ABC1 family protein |
| M. smegmatis MC2 155 | MSMEG_1353 | - | 0.0 | 84.65% (443) | ABC1 family protein |
| M. thermoresistible (build 8) | TH_3124 | - | 0.0 | 82.79% (459) | PUTATIVE ABC-1 domain protein |
| M. ulcerans Agy99 | MUL_0734 | - | 0.0 | 99.13% (461) | hypothetical protein MUL_0734 |
| M. vanbaalenii PYR-1 | Mvan_1241 | - | 0.0 | 86.52% (445) | hypothetical protein Mvan_1241 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5111|M.gilvum_PYR-GCK ----------------------------------------MSSTQRNQRT
Mvan_1241|M.vanbaalenii_PYR-1 ----------------------------------------MTAP-RN--T
MSMEG_1353|M.smegmatis_MC2_155 ----------------------------------------MSTTPAK--T
TH_3124|M.thermoresistible__bu ----------------------------------------MSTT------
MMAR_0982|M.marinum_M ----------------------------------------MTTT------
MUL_0734|M.ulcerans_Agy99 ------------------------------MTLSIWHTAQMTTT------
Mb0666c|M.bovis_AF2122/97 MRAEIGPDFRPHYTFGDAYPASERAHVNWELSAPVWHTAQMGST------
Rv0647c|M.tuberculosis_H37Rv MRAEIGPDFRPHYTFGDAYPASERAHVNWELSAPVWHTAQMGST------
MAV_4514|M.avium_104 --------------------------------------------------
MLBr_01898|M.leprae_Br4923 ----------------------------------------MSST------
MAB_3879|M.abscessus_ATCC_1997 --------------------------MTAQERSGRSAGEGRGAAGPAASS
Mflv_5111|M.gilvum_PYR-GCK PHRAVARLDRVPLPMEAARIGVTGWQITRTGGRVLSSLTTRG---TLQQK
Mvan_1241|M.vanbaalenii_PYR-1 PHRDVARLDRVPIPAEAARIGATGWQITRTGARVVTKLAGRG---SLQQK
MSMEG_1353|M.smegmatis_MC2_155 KHREVARLDRVPLPAEAARIGATGWQITRTGARVAANIFGRG---SLQQK
TH_3124|M.thermoresistible__bu KHREVAKLDRVSLPVEAARIGATGWQIARTGARVVTRLPSRG---SLQQK
MMAR_0982|M.marinum_M KHREVAKLDRVPLPVEAARVAVTGWQVTRTAARFVTKLPGKGFASSWQQK
MUL_0734|M.ulcerans_Agy99 KHREVAKLDRVPLPVEAARVAVTGWQVTRTAARFVTKLPGKGFASSWQQK
Mb0666c|M.bovis_AF2122/97 THREVAKLDRVPLPVEAARVAATGWQVTRTAVRFIGRLPRKG---PWQQK
Rv0647c|M.tuberculosis_H37Rv THREVAKLDRVPLPVEAARVAATGWQVTRTAVRFIGRLPRKG---PWQQK
MAV_4514|M.avium_104 ----MAKLDRVPLPVEAARVAATGWQVTRSAARVLTKLPAKG---PWQQK
MLBr_01898|M.leprae_Br4923 NNHQVVKLDRVPLPVEAARIAATYWQITRAAARVATKLAGKG---PWQQK
MAB_3879|M.abscessus_ATCC_1997 RGRQVAPLGRVALLSEAARLGTTGWQVARTAGRVLGKLTKKG---SLEQK
:. *.**.: ****:..* **::*:. *. : :* . :**
Mflv_5111|M.gilvum_PYR-GCK VVKQIPQAFADLGPTYVKFGQIIASSPGAFGEPLSREFRGLLDRVPPADS
Mvan_1241|M.vanbaalenii_PYR-1 IVKEIPQAFADLGPTYVKFGQIIASSPGAFGEPLSREFRSLLDRVPPADT
MSMEG_1353|M.smegmatis_MC2_155 IVKQVPKTFSDLGPTYVKFGQIIASSPGAFGEPLSREFRGLLDRVPPANP
TH_3124|M.thermoresistible__bu VIKELPKTFSDLGPTYVKFGQIIASSPGAFGEPLSREFRSLLDRVPPADP
MMAR_0982|M.marinum_M IIKEIPQTFADLGPTYVKFGQIIASSPGAFGESLSREFRGLLDRVPPADT
MUL_0734|M.ulcerans_Agy99 IIKEIPQTFVDLGPTYVKFGQIIASSPGAFGESLSREFRGLLDRVPPADT
Mb0666c|M.bovis_AF2122/97 VIKELPQTFADLGPTYVKFGQIIASSPGAFGESLSREFRGLLDRVPPAKT
Rv0647c|M.tuberculosis_H37Rv VIKELPQTFADLGPTYVKFGQIIASSPGAFGESLSREFRGLLDRVPPAKT
MAV_4514|M.avium_104 VIKQLPQTFADLGPTYVKFGQIIASSPGAFGESLSREFRGLLDRVPPADP
MLBr_01898|M.leprae_Br4923 VIREIPQTFANLGPTYVKFGQIIASSPSAFGESLAREFRGLLDQVPPADT
MAB_3879|M.abscessus_ATCC_1997 FIAELPQTFSDLGPTYVKLGQIIASSPGAFGEPLSREFRGLLDRVPPADP
.: ::*::* :*******:********.****.*:****.***:****..
Mflv_5111|M.gilvum_PYR-GCK DAVHKLLREDLGDEPQNLFKSFDEKPFASASIAQVHYATLHSGEEVVVKI
Mvan_1241|M.vanbaalenii_PYR-1 TAVHKLFREELGDEPQNLFKSFDDKPFASASIAQVHYATLHSGEEVVVKI
MSMEG_1353|M.smegmatis_MC2_155 DEVQKLIREELGDDPHKLFKTFDDTPFASASIAQVHYATLHSGEEVVVKI
TH_3124|M.thermoresistible__bu TAVRKLVRQELGADPSELFKHFDDKPFASASIAQVHYATLHSGEEVVVKI
MMAR_0982|M.marinum_M DQVHKLFVEELGAEPAELFAAFEEEPFASASIAQVHYATLKTGEEVVVKI
MUL_0734|M.ulcerans_Agy99 DQVHKLFVEELGTEPAELFATFEEEPFASASIAQVHYATLKTGEEVVVKI
Mb0666c|M.bovis_AF2122/97 DEVHKLFVEELGDEPARLFASFEEEPFASASIAQVHYATLRSGEEVVVKI
Rv0647c|M.tuberculosis_H37Rv DEVHKLFVEELGDEPARLFASFEEEPFASASIAQVHYATLRSGEEVVVKI
MAV_4514|M.avium_104 DEVHKLFLEELGADPSELFASWDETPFASASIAQVHYATLHTGEEVVVKI
MLBr_01898|M.leprae_Br4923 NEVHKLFVEELGDAPSKLFASFSEEPFASASIAQVHSAILHSGEHVVVKI
MAB_3879|M.abscessus_ATCC_1997 AAVRELIIAELGSEPEKLFASFDTEPFASASIAQVHYATLHSGEEVVVKI
*::*. :** * .** :. *********** * *::**.*****
Mflv_5111|M.gilvum_PYR-GCK QRPGIRRRVAADLQILKRGARLVEFAKLGQRLSAQDVVADFADNLAEELD
Mvan_1241|M.vanbaalenii_PYR-1 QRPGIRRRVAADLQILKRGARLVEFAKLGQRLSAQDVVADFADNLAEELD
MSMEG_1353|M.smegmatis_MC2_155 QRPGIRRRVAADLQILKRGARLVEFAKLGQRLSAQDVVADFADNLAEELD
TH_3124|M.thermoresistible__bu QRPGIRRRVTADLQILKHGARLVELAKLGRRLSAQDVVADFADNLAEELD
MMAR_0982|M.marinum_M QRPGIRRRVAADLQILQRFAQAVELAKLGRRLSAQDVVADFSDNLAEELD
MUL_0734|M.ulcerans_Agy99 QRPGIRRRVAADLQILQRFAQAVELAKLGRRLSAQDVVADFSDNLAVELD
Mb0666c|M.bovis_AF2122/97 QRPGIRRRVAADLQILKRFAQTVELAKLGRRLSAQDVVADFADNLAEELD
Rv0647c|M.tuberculosis_H37Rv QRPGIRRRVAADLQILKRFAQTVELAKLGRRLSAQDVVADFADNLAEELD
MAV_4514|M.avium_104 QRPGIRRRVAADLQILKRFAQAVELAKLGRRLSAQDVVADFSDNLAEELN
MLBr_01898|M.leprae_Br4923 QRPGIRRRVAADLQILKRFAQAVELAKLGRRLSAQDVVADFSDNLAEELN
MAB_3879|M.abscessus_ATCC_1997 QRPGIRKRVAADLQIMKRGAALLELGKVGRMLSARDVVDDFSGNLSEELD
******:**:*****::: * :*:.*:*: ***:*** **:.**: **:
Mflv_5111|M.gilvum_PYR-GCK FRLEAQSMDAWVAHMHSSPLGQNIRVPQVYWDLTSERVLTMERITGVRID
Mvan_1241|M.vanbaalenii_PYR-1 FRLEAQSMDAWVAHMHASPLGRNIRVPQVYWDLTSERVLTMERVTGVRID
MSMEG_1353|M.smegmatis_MC2_155 FRLEAQSMDAWVAHMHASPLGANIRVPTVYWDLTSERVLTMERITGVRID
TH_3124|M.thermoresistible__bu FRLEAQSMDTWVAHMHASPLGKNIRVPQVYWDLTSERVLTMERVSGVRID
MMAR_0982|M.marinum_M FRLEAQSMDAWVAHLHASPLGRNIRVPQVHWNFTSERVLTMERVHGVRID
MUL_0734|M.ulcerans_Agy99 FRLEAQSMDAWVAHLHASPLGRNIRVPQVHWNFTSERVLTMERVHGVRID
Mb0666c|M.bovis_AF2122/97 FRLEAQSMEAWVSHLHASPLGKNIRVPQVHWDFTTERVLTMERVHGIRID
Rv0647c|M.tuberculosis_H37Rv FRLEAQSMEAWVSHLHASPLGKNIRVPQVHWDFTTERVLTMERVHGIRID
MAV_4514|M.avium_104 FRLEAQSMEAWVSHLHASPLGKNIRVPQVYWDFTTDRVLTMERVSGIRID
MLBr_01898|M.leprae_Br4923 FRLEAQSMEAWGSYLHTSPLGKNIRVPKVHWEFTGERVLTMEQVHGIRID
MAB_3879|M.abscessus_ATCC_1997 FLAEGQAMQTWVEHLHTSSLGKNIRVPDVHWHYTTSKVLTMERVHGIRID
* *.*:*::* ::*:*.** ***** *:*. * .:*****:: *:***
Mflv_5111|M.gilvum_PYR-GCK DVAAIRKKGFDGEALVKALLFSVFEGGLRHGLFHGDLHAGNLYVDDDGKV
Mvan_1241|M.vanbaalenii_PYR-1 DVAAIRKKGFDGTELVKALLFSVFEGGLRHGLFHGDLHAGNLYVDDDGKI
MSMEG_1353|M.smegmatis_MC2_155 DVAAIRKKGFDGTELVKALLFSVFEGGLRHGLFHGDLHAGNLYVDDDGKI
TH_3124|M.thermoresistible__bu DVAALRKKGFEGTELVKALLFSLFEGGLRHGLFHGDLHAGNLYVDDDGKI
MMAR_0982|M.marinum_M NVAAIRKAGFDGTELVKALLFSVFEGGLRHGLFHGDLHAGNLYVDEEGRI
MUL_0734|M.ulcerans_Agy99 NVAAIRKAGFDGTELVKALLFSVFEGGLRHGLFHGDLHAGNLYVDEEGRI
Mb0666c|M.bovis_AF2122/97 NTAAIRKAGFDGVELVKALLFSVFEGGLRHGLFHGDLHAGNLYVDEAGRI
Rv0647c|M.tuberculosis_H37Rv NAAAIRKAGFDGVELVKALLFSVFEGGLRHGLFHGDLHAGNLYVDEAGRI
MAV_4514|M.avium_104 DVAAVRKAGFDGVELVKALLFSLFEGGLRHGLFHGDLHAGNLYVDEAGRI
MLBr_01898|M.leprae_Br4923 NADAIRKSGFDGVELVKALLFSVFEGGLRHGLFHGDLHAGNLHVDDQGRI
MAB_3879|M.abscessus_ATCC_1997 DAPAIRKAGFDGTELVKSLLFSVFESGLRQGLFHGDLHAGNLLVDDEGRI
:. *:** **:* ***:****:**.***:************ **: *::
Mflv_5111|M.gilvum_PYR-GCK VFFDFGIMGRIDPRTRWLLRELVYALLVKKDHAAAGKIVVLMGAVGTMRP
Mvan_1241|M.vanbaalenii_PYR-1 VFFDFGIMGRIDPRTRWLLRELVYALLVKKDHAAAGKIVVLMGAVGTMKP
MSMEG_1353|M.smegmatis_MC2_155 VFFDFGIMGRIDPRTRWLLRELVHALLVKKDHAAAGKIVVLMGAVGTVKP
TH_3124|M.thermoresistible__bu VFFDFGIMGRIDPRTRWLLRELVYALLVKKDHAAAGKIVVLMGAVGTTKP
MMAR_0982|M.marinum_M VFFDFGIMGRIDPRTRWLLRELVYALLVKKDHAAAGKIVVLMGAVGTMKP
MUL_0734|M.ulcerans_Agy99 VFFDFGIMGRIDPRTRWLLRELVYALLVKKDHAAAGKIVVLMGAVGTMKP
Mb0666c|M.bovis_AF2122/97 VFFDFGIMGRIDPRTRWLLRELVYALLVKKDHAAAGKIVVLMGAVGTMKP
Rv0647c|M.tuberculosis_H37Rv VFFDFGIMGRIDPRTRWLLRELVYALLVKKDHAAAGKIVVLMGAVGTMKP
MAV_4514|M.avium_104 VFFDFGIMGRIDPRTRWLLRELVYALLVKKDHAAAGKIVVLMGAVGTVKP
MLBr_01898|M.leprae_Br4923 VFFDFGIMGRIDPRTRWLLRELVFALLVKKDHAAAGKIVVLMGAVGTVKP
MAB_3879|M.abscessus_ATCC_1997 VFLDFGIVGFIDPRTRWLLRELIHALLVKKDHRAAGKIVVMLGAVGNPGD
**:****:* ************:.******** *******::****.
Mflv_5111|M.gilvum_PYR-GCK EAEAARDLEKFATPLTLKSLGDLSYAEIGKQLSTLADAYDVKLPRELVLI
Mvan_1241|M.vanbaalenii_PYR-1 EAEAAKDLEKFATPLTLKSLGDLSYAEIGKQLSALADAYDVKLPRELVLI
MSMEG_1353|M.smegmatis_MC2_155 EGQAAKDLEAFATPLTMKSLGDMSYAEIGRQLSALADAYDVKLPRELVLI
TH_3124|M.thermoresistible__bu EAQAAKDLEKFATPLTMSTLGDMSYAEIGRQLSALAEAYDVKLPRELVLI
MMAR_0982|M.marinum_M EAQAAKDLEKFATPLTMQSLGDMSYADIGRQLSALADAYDVKLPRELVLI
MUL_0734|M.ulcerans_Agy99 EAQAAKDLEKFATPLTMQSLGDMSYADIGRQLSALADAYDVKLPRELVLI
Mb0666c|M.bovis_AF2122/97 ETQAAKDLERFATPLTMQSLGDMSYADIGRQLSALADAYDVKLPRELVLI
Rv0647c|M.tuberculosis_H37Rv ETQAAKDLERFATPLTMQSLGDMSYADIGRQLSALADAYDVKLPRELVLI
MAV_4514|M.avium_104 EAQAAKDLEKFATPLTMQTLGDMSYADIGRQLSALADAYDVKLPRELVLI
MLBr_01898|M.leprae_Br4923 EARAAKDLENFTTPLAMKTLGDMSYTDIGRQLSALSDAYNVKLPRELVLI
MAB_3879|M.abscessus_ATCC_1997 AEQGTQDIQAFATPLTMKSLGDMSYSEIGKQLSTLAEQYDVKLPRELVLI
..::*:: *:***::.:***:**::**:***:*:: *:**********
Mflv_5111|M.gilvum_PYR-GCK GKQFLYVERYMKLLAPKWQMMSDPALTGYFANFMVEVSREHS-------D
Mvan_1241|M.vanbaalenii_PYR-1 GKQFLYVERYMKLLAPKWQMMSDPALTGYFANFMVEVSREHS-------D
MSMEG_1353|M.smegmatis_MC2_155 GKQFLYVERYMKLLAPKWQMMSDPQLTGYFANFMVEVSREHK-------E
TH_3124|M.thermoresistible__bu GKQFLYVERYMKLLAPKWQMMSDPELTGYFANFMVDIKREHD-------A
MMAR_0982|M.marinum_M GKQFLYVERYMKLLAPKWQMMSDPQLTGYFANFMVEVSREHQGDSSDASG
MUL_0734|M.ulcerans_Agy99 GKQFLYVERYMKLLAPKWQMMSDPQLTGYFANFMVEVSREHQGDSSDASG
Mb0666c|M.bovis_AF2122/97 GKQFLYVERYMKLLAPRWQMMSDPQLTGYFANFMVEVSREH---------
Rv0647c|M.tuberculosis_H37Rv GKQFLYVERYMKLLAPRWQMMSDPQLTGYFANFMVEVSREH---------
MAV_4514|M.avium_104 GKQFLYVERYMKLLAPKWQMMSDPQLTGYFANFMVEVSREHSSEF-----
MLBr_01898|M.leprae_Br4923 GKQFLYVERYMKLLAPEWQIMLDPELTGYFANFMVEVSREH---------
MAB_3879|M.abscessus_ATCC_1997 GKQFLYVERYMKLLAPRWQMMDDPELKGYFGNFIVDISREHN--------
****************.**:* ** *.***.**:*::.***
Mflv_5111|M.gilvum_PYR-GCK TGPDA--------------------
Mvan_1241|M.vanbaalenii_PYR-1 DVEA---------------------
MSMEG_1353|M.smegmatis_MC2_155 IDDVDEIPDIPETGTETQAPSGDKA
TH_3124|M.thermoresistible__bu KADELEV------------------
MMAR_0982|M.marinum_M DASDVEA------------------
MUL_0734|M.ulcerans_Agy99 DASDVEA------------------
Mb0666c|M.bovis_AF2122/97 -QSDIEV------------------
Rv0647c|M.tuberculosis_H37Rv -QSDIEV------------------
MAV_4514|M.avium_104 -ESDVEV------------------
MLBr_01898|M.leprae_Br4923 -QSDVEI------------------
MAB_3879|M.abscessus_ATCC_1997 REGDI--------------------