For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIRAVWNGTVLAETPRTARVEGNHYFPPESVHREYLRESSTTSLCPWKGVAHYYTVVVDGRTNPDAAWFY PRPSPLARRIKNHVAFWNGVDVEGEPEGEATGLVSRVAGWLGGKR
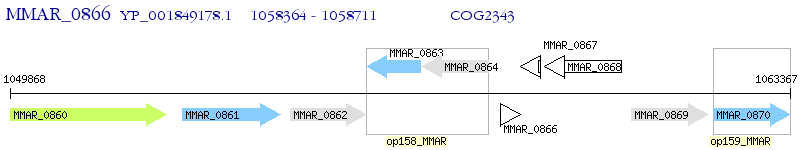
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0866 | - | - | 100% (115) | hypothetical protein MMAR_0866 |
| M. marinum M | MMAR_0350 | - | 7e-10 | 36.78% (87) | hypothetical protein MMAR_0350 |
| M. marinum M | MMAR_4414 | - | 2e-09 | 35.48% (93) | hypothetical protein MMAR_4414 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1697 | - | 1e-48 | 76.52% (115) | hypothetical protein Mb1697 |
| M. gilvum PYR-GCK | Mflv_4092 | - | 9e-10 | 39.74% (78) | hypothetical protein Mflv_4092 |
| M. tuberculosis H37Rv | Rv1670 | - | 1e-48 | 76.52% (115) | hypothetical protein Rv1670 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4612 | - | 1e-10 | 42.19% (64) | hypothetical protein MAB_4612 |
| M. avium 104 | MAV_5128 | - | 3e-09 | 35.56% (90) | hypothetical protein MAV_5128 |
| M. smegmatis MC2 155 | MSMEG_6469 | - | 6e-10 | 37.78% (90) | hypothetical protein MSMEG_6469 |
| M. thermoresistible (build 8) | TH_3885 | - | 5e-11 | 38.89% (90) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_0619 | - | 7e-66 | 97.39% (115) | hypothetical protein MUL_0619 |
| M. vanbaalenii PYR-1 | Mvan_5702 | - | 3e-12 | 39.53% (86) | hypothetical protein Mvan_5702 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6469|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5702|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4612|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4092|M.gilvum_PYR-GCK -----------MVATAHRREPRTDEPDENCCHRDTSMSLVAGRGPLSRDR
MAV_5128|M.avium_104 ------------------------------------MSLVAGRGPLSSDP
TH_3885|M.thermoresistible__bu MSTQVESAWPSHPDYRIDIEPFTYTAQVWFGDLLLAESDRCLILKEAKHI
MMAR_0866|M.marinum_M --------------------------------------------------
MUL_0619|M.ulcerans_Agy99 --------------------------------------------------
Mb1697|M.bovis_AF2122/97 --------------------------------------------------
Rv1670|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6469|M.smegmatis_MC2_155 -------------------------------------------------M
Mvan_5702|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4612|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4092|M.gilvum_PYR-GCK AGQLIPPVADDLVYVEPHPRRVQAVLGGRTVIDTEHALMVHRRGAPLSYA
MAV_5128|M.avium_104 AGRFSPPIPAEVVYVEPHPRRVQAVKDGRSVIDTERALMVHRRGRPLSYV
TH_3885|M.thermoresistible__bu DRLYFPDADIKWELLEQTDHTTVCPFKGRASYWTASHDGQTAENVAWSYR
MMAR_0866|M.marinum_M --------------------------------------------------
MUL_0619|M.ulcerans_Agy99 --------------------------------------------------
Mb1697|M.bovis_AF2122/97 --------------------------------------------------
Rv1670|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6469|M.smegmatis_MC2_155 NRKIGEARHVGPDMTSRP--------------------------------
Mvan_5702|M.vanbaalenii_PYR-1 -------------MTDRP--------------------------------
MAB_4612|M.abscessus_ATCC_1997 -----------MSIPDRP--------------------------------
Mflv_4092|M.gilvum_PYR-GCK FSAGDIGDLPSTPVPEAPGYAVVPWSAVDS------------WFEEGREL
MAV_5128|M.avium_104 FPADEVAGLPGEPEPEAPGFVHVPWDAVDT------------WWEEGRKL
TH_3885|M.thermoresistible__bu DPLPEVAAIAGHVAFYSERLRVVVVDRWPDGTVVHTRFPLWGDAAELLRL
MMAR_0866|M.marinum_M --------------------------------------------------
MUL_0619|M.ulcerans_Agy99 --------------------------------------------------
Mb1697|M.bovis_AF2122/97 --------------------------------------------------
Rv1670|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6469|M.smegmatis_MC2_155 -VLEPTAEHPITVTPTGRHVTVRVNGTVIAETDDALTLKEAS--------
Mvan_5702|M.vanbaalenii_PYR-1 -VLEPSPAHPITVVPTGRHVVVRVNGETIAETDSALTLQESS--------
MAB_4612|M.abscessus_ATCC_1997 -HLVPGPDHPIGISPSNTRVVVTAGDDTVADSTRALRLQEAS--------
Mflv_4092|M.gilvum_PYR-GCK VHYPPNPYHRVDCRPTARRLRVDAGEVTLVDTTDTTILFETA--------
MAV_5128|M.avium_104 VHYPPNPYHRVDCRGTRRRLRVRVGGTTLVDTDHTTIVFETA--------
TH_3885|M.thermoresistible__bu IDVVPEGDHFVGPAHGPTQRDVVEGGQLLAEAIVAVSKTLPDQRVTSASM
MMAR_0866|M.marinum_M ------------------MIRAVWNGTVLAETPRTARVEGNH--------
MUL_0619|M.ulcerans_Agy99 ------------------MIRAVWNGTVLAETPRTARVEGNH--------
Mb1697|M.bovis_AF2122/97 ------------------MIRAVWNGTVLAEAPRTVRVEGNH--------
Rv1670|M.tuberculosis_H37Rv ------------------MIRAVWNGTVLAEAPRTVRVEGNH--------
. . :.:: :
MSMEG_6469|M.smegmatis_MC2_155 --YPPVQYIPLADVDGSVLRRSDTTTYCPYKGEANYYSLEFGSGSEGAAL
Mvan_5702|M.vanbaalenii_PYR-1 --YPAVQYIPMADVVTERLRPSDTATYCPYKGQAGYFDVVAG----GQTV
MAB_4612|M.abscessus_ATCC_1997 --YPAVHYLPPDSVNWDVLQRTDTHTYCPYKGEASYYSVRTP----EGTV
Mflv_4092|M.gilvum_PYR-GCK --LPPRLYVRPSAVRTDLLRRSSTTSYCNYKGYATYWSLDID----GTVV
MAV_5128|M.avium_104 --LPPRLYVDPAHVRTDLLRRSETTSYCNYKGFATYWSLVDG----DRVV
TH_3885|M.thermoresistible__bu VFCKAASFTKPVDVSVDLLRGGRTFSTAEVR-ATQDGVLRGAAILLCDAG
MMAR_0866|M.marinum_M -------YFPPESVHREYLRESSTTSLCPWKGVAHYYTVVVDG----RTN
MUL_0619|M.ulcerans_Agy99 -------YFPPESVHRVYLRESTTTSLCPWKGVAHYYTVVVDG----RTN
Mb1697|M.bovis_AF2122/97 -------YFPPESLHREHLIESPTTSICPWKGLAHYYNVVVDGPY-GPVN
Rv1670|M.tuberculosis_H37Rv -------YFPPESLHREHLIESPTTSICPWKGLAHYYNVVVDGPY-GPVN
: : * * : . : : : .
MSMEG_6469|M.smegmatis_MC2_155 DDAIWTYEQPYPAVADIAGRVAFY-----PDKAEITVA------------
Mvan_5702|M.vanbaalenii_PYR-1 ADAIWTYREPHPAVAQIAGHVAFY-----ADKADVTVHP-----------
MAB_4612|M.abscessus_ATCC_1997 EDAAWTYEDPFPEVAPIARYLAFY-----PDRVTVTAE------------
Mflv_4092|M.gilvum_PYR-GCK DDVAWSYADPPPESLPIAGMVSFD-----ETRVAVTAELPATPERG----
MAV_5128|M.avium_104 DDVGWCYPDPPPESLPIKGFLSFD-----ETRVELLAELPVSARS-----
TH_3885|M.thermoresistible__bu ADDVFTHVDPMPEVPPPAGAVPFEG-FGMPGREIRVVDAAYDPDPDRVGP
MMAR_0866|M.marinum_M PDAAWFYPRPSPLARRIKNHVAFWNGVDVEGEPEGEATGLVSRVAGWLGG
MUL_0619|M.ulcerans_Agy99 PDAAWFYPRPSPLACRIKNHVAFWNGVDVEGEPEGEATGLVSRVAGWLGG
Mb1697|M.bovis_AF2122/97 PDAAWYYRRPSPLARRIKNHVAFWHGVTVEGESES-RHGLARRVVAWLG-
Rv1670|M.tuberculosis_H37Rv PDAAWYYRRPSPLARRIKNHVAFWHGVTVEGESES-RHGLARRVVAWLG-
* : : * * :.* .
MSMEG_6469|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5702|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4612|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4092|M.gilvum_PYR-GCK --------------------------------------------------
MAV_5128|M.avium_104 --------------------------------------------------
TH_3885|M.thermoresistible__bu PEINVWVRFRDAPPHQYLHTALLAQSTTHWTIAAGMLPHAGFGEAHAHRT
MMAR_0866|M.marinum_M KR------------------------------------------------
MUL_0619|M.ulcerans_Agy99 KR------------------------------------------------
Mb1697|M.bovis_AF2122/97 K-------------------------------------------------
Rv1670|M.tuberculosis_H37Rv K-------------------------------------------------
MSMEG_6469|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5702|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4612|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_4092|M.gilvum_PYR-GCK --------------------------------------------------
MAV_5128|M.avium_104 --------------------------------------------------
TH_3885|M.thermoresistible__bu LSTGIMKATVAFHDDADVTDWLLYHNRAFWAGRGLVQGEGRVFTQDGRQV
MMAR_0866|M.marinum_M --------------------------------------------------
MUL_0619|M.ulcerans_Agy99 --------------------------------------------------
Mb1697|M.bovis_AF2122/97 --------------------------------------------------
Rv1670|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6469|M.smegmatis_MC2_155 ---------------------------
Mvan_5702|M.vanbaalenii_PYR-1 ---------------------------
MAB_4612|M.abscessus_ATCC_1997 ---------------------------
Mflv_4092|M.gilvum_PYR-GCK ---------------------------
MAV_5128|M.avium_104 ---------------------------
TH_3885|M.thermoresistible__bu ASYTVQAMVRGFQRSPQEMGHDDRTAM
MMAR_0866|M.marinum_M ---------------------------
MUL_0619|M.ulcerans_Agy99 ---------------------------
Mb1697|M.bovis_AF2122/97 ---------------------------
Rv1670|M.tuberculosis_H37Rv ---------------------------