For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NRKIGEARHVGPDMTSRPVLEPTAEHPITVTPTGRHVTVRVNGTVIAETDDALTLKEASYPPVQYIPLAD VDGSVLRRSDTTTYCPYKGEANYYSLEFGSGSEGAALDDAIWTYEQPYPAVADIAGRVAFYPDKAEITVA *
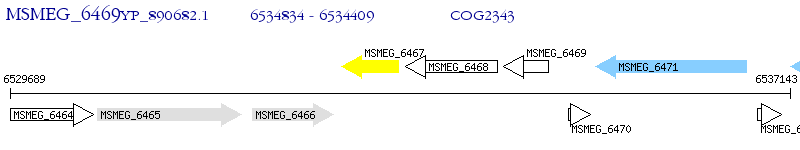
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6469 | - | - | 100% (141) | hypothetical protein MSMEG_6469 |
| M. smegmatis MC2 155 | MSMEG_2573 | - | 7e-15 | 32.76% (116) | hypothetical protein MSMEG_2573 |
| M. smegmatis MC2 155 | MSMEG_6499 | - | 1e-13 | 30.40% (125) | hypothetical protein MSMEG_6499 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0145 | - | 4e-41 | 62.70% (126) | hypothetical protein Mb0145 |
| M. gilvum PYR-GCK | Mflv_1110 | - | 2e-44 | 63.78% (127) | hypothetical protein Mflv_1110 |
| M. tuberculosis H37Rv | Rv0140 | - | 4e-41 | 62.70% (126) | hypothetical protein Rv0140 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4612 | - | 8e-30 | 48.36% (122) | hypothetical protein MAB_4612 |
| M. marinum M | MMAR_0350 | - | 3e-38 | 60.32% (126) | hypothetical protein MMAR_0350 |
| M. avium 104 | MAV_5156 | - | 1e-37 | 58.27% (127) | hypothetical protein MAV_5156 |
| M. thermoresistible (build 8) | TH_2320 | - | 5e-18 | 38.40% (125) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_4770 | - | 2e-38 | 60.32% (126) | hypothetical protein MUL_4770 |
| M. vanbaalenii PYR-1 | Mvan_5702 | - | 2e-44 | 66.67% (126) | hypothetical protein Mvan_5702 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1110|M.gilvum_PYR-GCK MN----------GDMTDRPIHEPSPS--HPITVVPTGRHVVVRVNGDVVA
Mvan_5702|M.vanbaalenii_PYR-1 --------------MTDRPVLEPSPA--HPITVVPTGRHVVVRVNGETIA
MSMEG_6469|M.smegmatis_MC2_155 MNRKIGEARHVGPDMTSRPVLEPTAE--HPITVTPTGRHVTVRVNGTVIA
Mb0145|M.bovis_AF2122/97 --------------MSNRIVLEPSAD--HPITIEPTNRRVQVRVNGEVVA
Rv0140|M.tuberculosis_H37Rv --------------MSNRIVLEPSAD--HPITIEPTNRRVQVRVNGEVVA
MMAR_0350|M.marinum_M --------------MSHRKVLEPSAG--HPITIEPTKGRVQVRVNGELIA
MUL_4770|M.ulcerans_Agy99 --------------MSHRKVLEPSAG--HPITIEPTKGRVQVRVNGELIA
MAV_5156|M.avium_104 --------------MAHPEIKEPSAG--HPITIEPTRGRVQVRVNGELIA
MAB_4612|M.abscessus_ATCC_1997 ------------MSIPDRPHLVPGPD--HPIGISPSNTRVVVTAGDDTVA
TH_2320|M.thermoresistible__bu ---VILDFGAFEWCEEDEPVVSHPRDPFHRIDVCRTSRPVRVELDGTVLA
* * : : * * .. :*
Mflv_1110|M.gilvum_PYR-GCK ETDAALTLQEASYPAVQYIPIGDVLPGRLSPSDTATYCPFKGDAGYFDVT
Mvan_5702|M.vanbaalenii_PYR-1 ETDSALTLQESSYPAVQYIPMADVVTERLRPSDTATYCPYKGQAGYFDVV
MSMEG_6469|M.smegmatis_MC2_155 ETDDALTLKEASYPPVQYIPLADVDGSVLRRSDTTTYCPYKGEANYYSLE
Mb0145|M.bovis_AF2122/97 DTAAALCLQEASYPAVQYIPLADVVQDRLIRTETSTYCPFKGEASYYSVT
Rv0140|M.tuberculosis_H37Rv DTAAALCLQEASYPAVQYIPLADVVQDRLIRTETSTYCPFKGEASYYSVT
MMAR_0350|M.marinum_M DTAAALELREATLPAVQYIPRADVVADRLTSTDTSSYCPFKGDASYYSVT
MUL_4770|M.ulcerans_Agy99 DTAAALELREATLPAVQYIPRADVVADRLTSTDTSSYCPFKGDASYYSVT
MAV_5156|M.avium_104 DSTAALELREATLPAVQYIPFTDVAQDRLTRTDTRTYCPFKGEASYYSVT
MAB_4612|M.abscessus_ATCC_1997 DSTRALRLQEASYPAVHYLPPDSVNWDVLQRTDTHTYCPYKGEASYYSVR
TH_2320|M.thermoresistible__bu ESDRAAVLFETGLPPRWYLPRDDVVAELLP-SDTVTYCAYKGRACYYSLP
:: * * *: *. *:* .* * ::* :**.:** * *:.:
Mflv_1110|M.gilvum_PYR-GCK AG----GSTVTDAVWTYREPYPAVAQIAGHVAFYPDKADVSVDDG-----
Mvan_5702|M.vanbaalenii_PYR-1 AG----GQTVADAIWTYREPHPAVAQIAGHVAFYADKADVTVHP------
MSMEG_6469|M.smegmatis_MC2_155 FGSGSEGAALDDAIWTYEQPYPAVADIAGRVAFYPDKAEITVA-------
Mb0145|M.bovis_AF2122/97 TD---AGDIVDDVMWTYENPYPAVAAIAGHVACYPDKAEISIFPG-----
Rv0140|M.tuberculosis_H37Rv TD---AGDIVDDVMWTYENPYPAVAAIAGHVACYPDKAEISIFPG-----
MMAR_0350|M.marinum_M TS---TGDTVADVIWTYEQPYPAVAEIAGHVAFYPNKAEISVIPD-----
MUL_4770|M.ulcerans_Agy99 TS---TGDTVADVIWTYEQPYPAVAEIAGHVAFYPNKAEISVIPD-----
MAV_5156|M.avium_104 TS---AGDTVDDVIWTYEQPYPAVAAIAGHAAFYPDKAEISISTD-----
MAB_4612|M.abscessus_ATCC_1997 TP----EGTVEDAAWTYEDPFPEVAPIARYLAFYPDRVTVTAE-------
TH_2320|M.thermoresistible__bu DG-------PADVAWSYREPLHDAVPVRDRICFFDERVDVVVDGQRRQRP
*. *:*.:* .. : . : ::. :
Mflv_1110|M.gilvum_PYR-GCK -----
Mvan_5702|M.vanbaalenii_PYR-1 -----
MSMEG_6469|M.smegmatis_MC2_155 -----
Mb0145|M.bovis_AF2122/97 -----
Rv0140|M.tuberculosis_H37Rv -----
MMAR_0350|M.marinum_M -----
MUL_4770|M.ulcerans_Agy99 -----
MAV_5156|M.avium_104 -----
MAB_4612|M.abscessus_ATCC_1997 -----
TH_2320|M.thermoresistible__bu TTVWS