For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVATAHRREPRTDEPDENCCHRDTSMSLVAGRGPLSRDRAGQLIPPVADDLVYVEPHPRRVQAVLGGRTV IDTEHALMVHRRGAPLSYAFSAGDIGDLPSTPVPEAPGYAVVPWSAVDSWFEEGRELVHYPPNPYHRVDC RPTARRLRVDAGEVTLVDTTDTTILFETALPPRLYVRPSAVRTDLLRRSSTTSYCNYKGYATYWSLDIDG TVVDDVAWSYADPPPESLPIAGMVSFDETRVAVTAELPATPERG
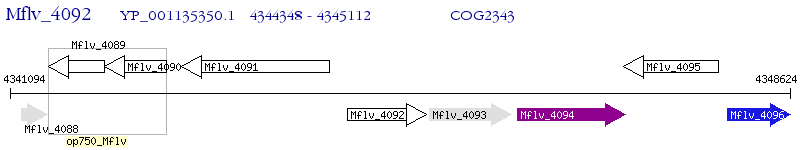
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4092 | - | - | 100% (254) | hypothetical protein Mflv_4092 |
| M. gilvum PYR-GCK | Mflv_1110 | - | 2e-11 | 29.91% (117) | hypothetical protein Mflv_1110 |
| M. gilvum PYR-GCK | Mflv_2745 | - | 8e-09 | 27.86% (140) | hypothetical protein Mflv_2745 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1085 | - | 1e-24 | 34.12% (211) | hypothetical protein Mb1085 |
| M. tuberculosis H37Rv | Rv1056 | - | 1e-24 | 34.12% (211) | hypothetical protein Rv1056 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4612 | - | 8e-21 | 41.53% (118) | hypothetical protein MAB_4612 |
| M. marinum M | MMAR_0394 | - | 1e-85 | 66.52% (224) | hypothetical protein MMAR_0394 |
| M. avium 104 | MAV_5128 | - | 1e-94 | 72.44% (225) | hypothetical protein MAV_5128 |
| M. smegmatis MC2 155 | MSMEG_2573 | - | 8e-87 | 66.07% (224) | hypothetical protein MSMEG_2573 |
| M. thermoresistible (build 8) | TH_0282 | - | 2e-88 | 67.26% (223) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1044 | - | 7e-85 | 66.07% (224) | hypothetical protein MUL_1044 |
| M. vanbaalenii PYR-1 | Mvan_2252 | - | 1e-101 | 76.00% (225) | hypothetical protein Mvan_2252 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4092|M.gilvum_PYR-GCK MVATAHRREPRTDEPDENCCHRDTSMSLVAGRGPLSRDRAGQLIPPVADD
Mvan_2252|M.vanbaalenii_PYR-1 -------------------------MSLVAGHGPLSRDRAGVLSPPVAGD
MMAR_0394|M.marinum_M -------------------------MSLAAGHGPLSTHPAGRFCPPVPDG
MUL_1044|M.ulcerans_Agy99 -------------------------MGLAAGHGPLSTHPAGRFCPPVPDG
TH_0282|M.thermoresistible__bu -------------------------VSLTAGHGPLGADPAGWFTPPIPDE
MAV_5128|M.avium_104 -------------------------MSLVAGRGPLSSDPAGRFSPPIPAE
MSMEG_2573|M.smegmatis_MC2_155 -------------------------MSLVAGHGPLSNRPAGWFSPPLPAP
MAB_4612|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1085|M.bovis_AF2122/97 ----MSVDYPQMAATRGRIEPAPRRVRGYLGHVLVFDTSAARYVWEVPYY
Rv1056|M.tuberculosis_H37Rv ----MSVDYPQMAATRGRIEPAPRRVRGYLGHVLVFDTSAARYVWEVPYY
Mflv_4092|M.gilvum_PYR-GCK LVYVEPHPRRVQAVLGGRTVIDTEHALMVHRRGAPLSYAFSAGDIGDLPS
Mvan_2252|M.vanbaalenii_PYR-1 LVYIEPHPRRIRAILDGRTVIDTETALMVHRRGSPLSYAFAAADVGDLPG
MMAR_0394|M.marinum_M LVYVEPHPRRIQAFRNGRLVIDTERALMVHRRGHPLRYVFPTDEIADLPT
MUL_1044|M.ulcerans_Agy99 LVYVEPHPRRIQAFRNGRLVIDTERALMVHRRGHPLRYVFPTDEITDLPT
TH_0282|M.thermoresistible__bu VVFVEPHPRRIQGVRDGRAVIDTERALLVHRRDRPLSYAFPADVVGDLPQ
MAV_5128|M.avium_104 VVYVEPHPRRVQAVKDGRSVIDTERALMVHRRGRPLSYVFPADEVAGLPG
MSMEG_2573|M.smegmatis_MC2_155 AVYIEPHPRRVQAFVGGDALIDTECALMVHRAGSPLSYAFPVDEVGGLPA
MAB_4612|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1085|M.bovis_AF2122/97 PQYYIPLADVRMEFLRDENHPQRVQLGPSRLHSLVSAGQTHRSAARVFDV
Rv1056|M.tuberculosis_H37Rv PQYYIPLADVRMEFLRDENHPQRVQLGPSRLHSLVSAGQTHRSAARVFDV
Mflv_4092|M.gilvum_PYR-GCK TPVPEAPGYAVVPWSAVDSWFEEGRELVHYPPNPYHRVDCRPTARRLRVD
Mvan_2252|M.vanbaalenii_PYR-1 EPVPEAPGFVVVPWAAVDAWWEEGRRLVHYPPNPYHRVDCRPTSRRLRVA
MMAR_0394|M.marinum_M QPETAAPGFVHVRWDAVDMWLEEGRRLVHYPPNPYHRVDCRPTTRQLHVE
MUL_1044|M.ulcerans_Agy99 QPETAAPGFVHVRWDAVDMWLEEGRRLVHYPPNPYHRVDCRPTTRQLHVE
TH_0282|M.thermoresistible__bu RPEPAAPGYVQVPWDAVEAWVEEGRRLVHYPPNPYHRVDCHPTRRALRVA
MAV_5128|M.avium_104 EPEPEAPGFVHVPWDAVDTWWEEGRKLVHYPPNPYHRVDCRGTRRRLRVR
MSMEG_2573|M.smegmatis_MC2_155 VAVPEAPGYVTVPWDAVEVWMEEGRRLVHYPPNPYHRIDCRPTKRRLRVA
MAB_4612|M.abscessus_ATCC_1997 MSIPDRP---------------------HLVPGPDHPIGISPSNTRVVVT
Mb1085|M.bovis_AF2122/97 DGDSPVAGTVRFNWDPLR-WFEEDEPIYGHPRNPYQRADALRSHRHVRVE
Rv1056|M.tuberculosis_H37Rv DGDSPVAGTVRFNWDPLR-WFEEDEPIYGHPRNPYQRADALRSHRHVRVE
. . .* : . : : *
Mflv_4092|M.gilvum_PYR-GCK AGEVTLVDTTDTTILFETALPPRLYVRPSAVRTDLLRRSSTTSYCNYKGY
Mvan_2252|M.vanbaalenii_PYR-1 VAGTPLVDTVDIVILFETALQPRLYVSPALVRTDLLRRSDTTSYCNYKGY
MMAR_0394|M.marinum_M LAGTTLVNTQDTVIVFETSLQPRLYVTPAQVRTDLLVRSQTSSYCNYKGV
MUL_1044|M.ulcerans_Agy99 LAGTTLVNTQDTVIVFETSLQPRLYVTPAQVRTDLLVRSQTSSYCNYKGV
TH_0282|M.thermoresistible__bu VEGTVLVDTTETVIVFETSLAPRLYVAPELVRTDLLRRTDTSSYCNYKGH
MAV_5128|M.avium_104 VGGTTLVDTDHTTIVFETALPPRLYVDPAHVRTDLLRRSETTSYCNYKGF
MSMEG_2573|M.smegmatis_MC2_155 VGDTVLVDTDDTIIVFETALEPRLYVSPALVRTDLLHRTDTTTYCNYKGV
MAB_4612|M.abscessus_ATCC_1997 AGDDTVADSTRALRLQEASYPAVHYLPPDSVNWDVLQRTDTHTYCPYKGE
Mb1085|M.bovis_AF2122/97 LDGIVLADTRSPVLLFETGIPTRYYIDPADIAFEHLEPTSTQTLCPYKGT
Rv1056|M.tuberculosis_H37Rv LDGIVLADTRSPVLLFETGIPTRYYIDPADIAFEHLEPTSTQTLCPYKGT
:.:: : *:. . *: * : : * :.* : * ***
Mflv_4092|M.gilvum_PYR-GCK AT-YWSLDIDGTVVDDVAWSYADPPPESLPIAGMVSFDETRVAVTAELPA
Mvan_2252|M.vanbaalenii_PYR-1 AT-YWSAVVGDTVVEDVAWSYPDPPPESLPIRDMLSFDPGKVDLTAELPG
MMAR_0394|M.marinum_M AT-YWSAVVGDMVAPDIAWSYEDPPPESVPIKGFLSFDEGAVQVTAELPA
MUL_1044|M.ulcerans_Agy99 AT-YWSAVVGDMVAPDIAWSYEDPPPESVPIKGFLSFDEGVVQVTAELPA
TH_0282|M.thermoresistible__bu AT-YWAAVIGDTVVEDVAWSYEDPRPESLPIKGYLSFDDTRVQMTAELPG
MAV_5128|M.avium_104 AT-YWSLVDGDRVVDDVGWCYPDPPPESLPIKGFLSFDETRVELLAELPV
MSMEG_2573|M.smegmatis_MC2_155 AT-YWAAVVADTVISDVAWSYPDTPPEAEPLRGHLSFDPARVDVVAELPA
MAB_4612|M.abscessus_ATCC_1997 AS-YYSVRTPEGTVEDAAWTYEDPFPEVAPIARYLAFYPDRVTVTAE---
Mb1085|M.bovis_AF2122/97 TSGYWSVRVGDAVHRDLAWTYHYPLPAVAPIAGLVAFYNEKVDLTVDGVA
Rv1056|M.tuberculosis_H37Rv TSGYWSVRVGDAVHRDLAWTYHYPLPAVAPIAGLVAFYNEKVDLTVDGVA
:: *:: . * .* * . * *: ::* * : .:
Mflv_4092|M.gilvum_PYR-GCK TPERG------
Mvan_2252|M.vanbaalenii_PYR-1 S----------
MMAR_0394|M.marinum_M R----------
MUL_1044|M.ulcerans_Agy99 R----------
TH_0282|M.thermoresistible__bu LS---------
MAV_5128|M.avium_104 SARS-------
MSMEG_2573|M.smegmatis_MC2_155 GASTAACGCSV
MAB_4612|M.abscessus_ATCC_1997 -----------
Mb1085|M.bovis_AF2122/97 LPRPHTQFS--
Rv1056|M.tuberculosis_H37Rv LPRPHTQFS--