For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPGQPTRDQVAALVDHTLLKPEATAADVVALVAEAADLGVYAVCVSPSMVPAAVSAGGVRVATVAGFPSG KHASAIKAHEAALAVACGAVEVDMVIDVGAALAGHLDAVRSDIEAVRCATSGAVLKVIVESAALLGLADE STLIGVCRVAEDAGADFVKTSTGFHPAGGASTRAVEVMASAVGGRLGVKASGGIRTATDAVAMLSAGATR LGLSGTRAVLEGLGQN
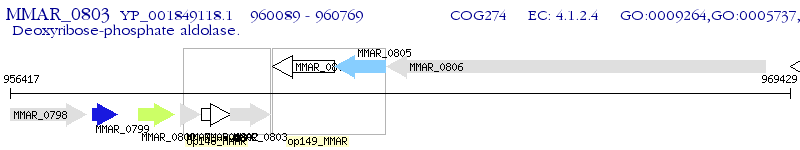
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0803 | deoC | - | 100% (226) | deoxyribose-phosphate aldolase DeoC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0488 | deoC | 1e-94 | 79.64% (221) | deoxyribose-phosphate aldolase |
| M. gilvum PYR-GCK | Mflv_0097 | - | 9e-80 | 71.62% (222) | deoxyribose-phosphate aldolase |
| M. tuberculosis H37Rv | Rv0478 | deoC | 1e-94 | 79.64% (221) | deoxyribose-phosphate aldolase |
| M. leprae Br4923 | MLBr_02451 | deoC | 1e-89 | 78.03% (223) | deoxyribose-phosphate aldolase |
| M. abscessus ATCC 19977 | MAB_4078c | - | 1e-74 | 69.30% (215) | deoxyribose-phosphate aldolase (DeoC) |
| M. avium 104 | MAV_4672 | deoC | 1e-96 | 83.26% (221) | deoxyribose-phosphate aldolase |
| M. smegmatis MC2 155 | MSMEG_0922 | deoC | 2e-81 | 73.30% (221) | deoxyribose-phosphate aldolase |
| M. thermoresistible (build 8) | TH_0872 | deoC | 2e-47 | 76.15% (130) | PROBABLE DEOXYRIBOSE-PHOSPHATE ALDOLASE DEOC |
| M. ulcerans Agy99 | MUL_4546 | deoC | 1e-121 | 100.00% (226) | deoxyribose-phosphate aldolase |
| M. vanbaalenii PYR-1 | Mvan_0811 | - | 3e-83 | 74.21% (221) | deoxyribose-phosphate aldolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0097|M.gilvum_PYR-GCK -MGSYTRAQVAALVDHTLLKPEATEADIEAVVAEALALEVKAICVSPTFV
Mvan_0811|M.vanbaalenii_PYR-1 -MGSYSRARVAALVDHTLLKPEATEADIEALVTEAAELGVYAICVSPTFV
MMAR_0803|M.marinum_M MPGQPTRDQVAALVDHTLLKPEATAADVVALVAEAADLGVYAVCVSPSMV
MUL_4546|M.ulcerans_Agy99 MPGQPTRDQVAALVDHTLLKPEATAADVVALVAEAADLGVYAVCVSPSMV
Mb0488|M.bovis_AF2122/97 MLGQPTRAQLAALVDHTLLKPETTRADVAALVAEAAELGVYAVCVSPSMV
Rv0478|M.tuberculosis_H37Rv MLGQPTRAQLAALVDHTLLKPETTRADVAALVAEAAELGVYAVCVSPSMV
MLBr_02451|M.leprae_Br4923 MQGQPTRKQLAALVDHTLLKSEATHADVAALVAEAVELGVYAVCVSPSMV
MAV_4672|M.avium_104 MT--PTRAQLAAFVDHTLLKPEATAADVAALVTEAAELGVYAVCVSPPMV
TH_0872|M.thermoresistible__bu --------------------------------------------------
MSMEG_0922|M.smegmatis_MC2_155 -MTDHTRAQVAALVDHTLLKPEATPSDVTALVDEAADLGVFAVCVSPPLV
MAB_4078c|M.abscessus_ATCC_199 ---------MAALIDHTLLKPEATPEDVRALVAEAVDLGVLAVCVSPSML
Mflv_0097|M.gilvum_PYR-GCK AAVASLAP-AATVASVVGFPSGKHLSSVKAAEAHAAAAAGAAEIDMVIDV
Mvan_0811|M.vanbaalenii_PYR-1 AAVSLLAP-DATVASVVGFPSGKHVSAIKAEEARLAVEAGAREVDMVIDV
MMAR_0803|M.marinum_M PAAVSAG--GVRVATVAGFPSGKHASAIKAHEAALAVACGAVEVDMVIDV
MUL_4546|M.ulcerans_Agy99 PAAVSAG--GVRVATVAGFPSGKHASAIKAHEAALAVACGAVEVDMVIDV
Mb0488|M.bovis_AF2122/97 PVAVQAG--GVRVAAVTGFPSGKHVSSVKAHEAAAALASGASEIDMVIDI
Rv0478|M.tuberculosis_H37Rv PVAVQAG--GVRVAAVTGFPSGKHVSSVKAHEAAAALASGASEIDMVIDI
MLBr_02451|M.leprae_Br4923 PAAVEAGAGGLRVATVVGFPSGEHVSIVKAQEAVQAVASGATEIDMVIAV
MAV_4672|M.avium_104 PAAVQAG-AGVRVASVAGFPSGKHVSAVKAHEAALAVASGAAEIDMVIDV
TH_0872|M.thermoresistible__bu ---------------------------------------------MVIDV
MSMEG_0922|M.smegmatis_MC2_155 SVAAGVAPSGLAIAAVAGFPSGKHVPGIKATEAELAVAAGATEIDMVIDV
MAB_4078c|M.abscessus_ATCC_199 PVLLPEGVERLLVAGVVGFPSGKHLSSVKAHEAALAVAAGAHEIDMVIDV
*** :
Mflv_0097|M.gilvum_PYR-GCK GAALAGQLDAVRADIAAVRAAAPETVLKVIVESAALLEFGGQPILLDVCR
Mvan_0811|M.vanbaalenii_PYR-1 GCALAGELGAVRSDIAAVRAAAPEAVLKVIVESAAILQISGEPLLVDICR
MMAR_0803|M.marinum_M GAALAGHLDAVRSDIEAVRCATSGAVLKVIVESAALLGLADESTLIGVCR
MUL_4546|M.ulcerans_Agy99 GAALAGHLDAVRSDIEAVRCATSGAVLKVIVESAALLGLADESTLIGVCR
Mb0488|M.bovis_AF2122/97 GAALCGDIDAVRSDIEAVRAAAAGAVLKVIVESAVLLGQSNAHTLVDACR
Rv0478|M.tuberculosis_H37Rv GAALCGDIDAVRSDIEAVRAAAAGAVLKVIVESAVLLGQSNAHTLVDACR
MLBr_02451|M.leprae_Br4923 GAALAGDLDAVRSDIEMVRSAIAGVVLKVIVESAVLLGQANERTLAEVCH
MAV_4672|M.avium_104 GAALAGDLDGVRADIAAVRGAVGGAVLKVIVESSALLALADEHTLVRVCR
TH_0872|M.thermoresistible__bu GAAVAGELGAVYDDIAEVRRAVSGITLKVIVESAVLLEHTDPERLTAVCR
MSMEG_0922|M.smegmatis_MC2_155 GAALAGDLDAVSADITAVRKAVRAATLKVIVESAALLEFSGEPLLADVCR
MAB_4078c|M.abscessus_ATCC_199 GAAVGGDFGAVQGDIEAVRRAVPAATLKVIIESAALLERSGPEAVRAACR
*.*: *.:..* ** ** * .****:**:.:* . : *:
Mflv_0097|M.gilvum_PYR-GCK IAEDAGAEFVKTSTGFHPSGGASPRAVELMAGAVGGRLGVKASGGIRTAA
Mvan_0811|M.vanbaalenii_PYR-1 VAEDAGADFVKTSTGFHPSGGASVRAVELMAGAVGGRLGVKASGGIRSAA
MMAR_0803|M.marinum_M VAEDAGADFVKTSTGFHPAGGASTRAVEVMASAVGGRLGVKASGGIRTAT
MUL_4546|M.ulcerans_Agy99 VAEDAGADFVKTSTGFHPAGGASTRAVEVMASAVGGRLGVKASGGIRTAT
Mb0488|M.bovis_AF2122/97 AAEDAGADFVKTSTGCHPAGGATVRAVELMAETVGPRLGVKASGGIRTAA
Rv0478|M.tuberculosis_H37Rv AAEDAGADFVKTSTGCHPAGGATVRAVELMAETVGPRLGVKASGGIRTAA
MLBr_02451|M.leprae_Br4923 VAEAAGADFVKTSTGFHPAGGASVRAVTLMTEAVGGRLGVKASGGIRTAV
MAV_4672|M.avium_104 AAEDAGADFVKTSTGFHPSGGASVRAVALMAEAVGGRLGVKASGGIRTAA
TH_0872|M.thermoresistible__bu AAEDAGADFVKTSTGFHPSGGASVQAVSVMAAAVGGRLGVKASGGIRTAA
MSMEG_0922|M.smegmatis_MC2_155 VARDAGADFVKTSTGFHPSGGASVQAVEIMARTVGERLGVKASGGIRTAE
MAB_4078c|M.abscessus_ATCC_199 ASEAAGADFVKTSTGFHPAGGATVEAVRIMAETVGDRLQVKASGGIRTAR
:. ***:******* **:***: .** :*: :** ** ********:*
Mflv_0097|M.gilvum_PYR-GCK AATEMLDAGATRLGLSGTRAVLDGLRA-
Mvan_0811|M.vanbaalenii_PYR-1 DAAAMLDAGATRLGLSGTRAVLAGFPD-
MMAR_0803|M.marinum_M DAVAMLSAGATRLGLSGTRAVLEGLGQN
MUL_4546|M.ulcerans_Agy99 DAVAMLSAGATRLGLSGTRAVLEGLGQN
Mb0488|M.bovis_AF2122/97 DAVAMLNAGATRLGLSGTRAVLDGLS--
Rv0478|M.tuberculosis_H37Rv DAVAMLNAGATRLGLSGTRAVLDGLS--
MLBr_02451|M.leprae_Br4923 DAIALLDAGATRLGLSSTREVLDGLS--
MAV_4672|M.avium_104 DALAMLDAGATRLGLSGTRAVLDGLG--
TH_0872|M.thermoresistible__bu DAVAMLDAGATRLGLSASRAVLDGLT--
MSMEG_0922|M.smegmatis_MC2_155 QAAAMLDAGATRLGLSGSRAVLDGFGSA
MAB_4078c|M.abscessus_ATCC_199 DAAAMLEAGATRLGLSGSRIVLDGFAD-
* :*.*********.:* ** *: