For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAKDDLDSNPGTEPFVPEFDTGERSVPFVPDFDTDSKPFAGLAAVVTESKKSSVELVDLSEVSAPLTTLD GDSLDGDSPAEGTATPAPSVTVPSRYVYLKWWKFVLVVLGVWFGAAQVGLSLFYWWFHTIDKTLPVFVVL VYLVLCAVASLLLSMVQGRPLITALSLALMSAPFASVAAAAPLYGSYFCEHASRCLAGVIPY
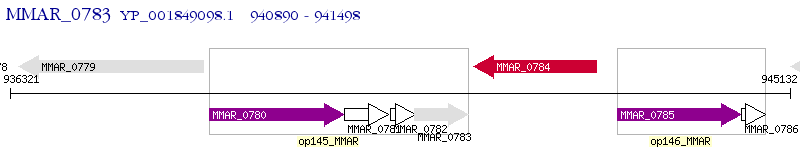
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0783 | - | - | 100% (202) | transmembrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0470 | - | 3e-52 | 60.11% (178) | hypothetical protein Mb0470 |
| M. gilvum PYR-GCK | Mflv_0120 | - | 7e-28 | 51.30% (115) | hypothetical protein Mflv_0120 |
| M. tuberculosis H37Rv | Rv0461 | - | 3e-52 | 60.11% (178) | hypothetical protein Rv0461 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4128c | - | 4e-18 | 32.63% (190) | hypothetical protein MAB_4128c |
| M. avium 104 | MAV_4688 | - | 8e-47 | 46.38% (207) | hypothetical protein MAV_4688 |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_2920 | - | 1e-29 | 40.78% (179) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1419 | - | 1e-104 | 93.07% (202) | transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_0793 | - | 1e-27 | 46.83% (126) | hypothetical protein Mvan_0793 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0783|M.marinum_M ---------------------------------------------MAKDD
MUL_1419|M.ulcerans_Agy99 ---------------------------------------------MAKDD
Mb0470|M.bovis_AF2122/97 --------------------------------------------------
Rv0461|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4688|M.avium_104 ---------------------------------------------MADDK
Mflv_0120|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0793|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2920|M.thermoresistible__bu ---------------------------------------------VPDRR
MAB_4128c|M.abscessus_ATCC_199 MSDKNAENTVEKTDAEADGAAAELVDDADQPSVVEGDLADPLAVPIDDGK
MMAR_0783|M.marinum_M LDSNPGTEPFVPEFDTGERSVPFVPDFDT--DSKPFAGLAAVVTESKKSS
MUL_1419|M.ulcerans_Agy99 LDSNPGTEP-------------FVPDFDM--DSKPFAGLAAVVTESKKSS
Mb0470|M.bovis_AF2122/97 -----------------------MPDFDTGAHSQRFLSLAGQQDRAGKSW
Rv0461|M.tuberculosis_H37Rv -----------------------MPDFDTGAHSQRFLSLAGQQDRAGKSW
MAV_4688|M.avium_104 DASSPGTEA-------------FVPDFSDDDDAADTGTQSWAPDFDDDAE
Mflv_0120|M.gilvum_PYR-GCK -----------------------------MS-EQTILEDPS---------
Mvan_0793|M.vanbaalenii_PYR-1 -----------------------------MSEDQIALDDPA---------
TH_2920|M.thermoresistible__bu SETLPGEPTG-----------QYTPVFDTGSFTAVQPDDPARVWEADPVK
MAB_4128c|M.abscessus_ATCC_199 ARDGLDTATFTAFSAFKDDEDDLVPTGRIDTDALPLTEAFDVGSDRTGAI
MMAR_0783|M.marinum_M VELVDLSEVSAPLTTLDGDSLDGDSPAEGTATPAPSVTVPS--RYVYLKW
MUL_1419|M.ulcerans_Agy99 VELVDLSEVSAPLTTLDGDSLDGDSPAEGTATPAPSVTVPS--RYVYLKW
Mb0470|M.bovis_AF2122/97 PGSTPKPQE-DPVGVAPSASVEVLG-SEPAATLAHSVTVPG--RYTYLKW
Rv0461|M.tuberculosis_H37Rv PGSTPKPQE-DPVGVAPSASVEVLG-SEPAATLAHSVTVPG--RYTYLKW
MAV_4688|M.avium_104 DDDTEAAEPAAERGPEPEPEPEPAG----FAGPVPPVAVPG--RYFYVKW
Mflv_0120|M.gilvum_PYR-GCK ------------------PSNELP------PPHAQPIVVPS--AEQSVSR
Mvan_0793|M.vanbaalenii_PYR-1 ------------------YADELP------RPPAQPVVVPA--AAQSVGR
TH_2920|M.thermoresistible__bu PLAAEFEPATDSPQTGDVPIGELPS--EPIPAPARPMVVPG--SYQFLKR
MAB_4128c|M.abscessus_ATCC_199 PAAVGRSRAEAMTPDPYTGAVPVLG-----DEPAKPVRVPAEREPALLKR
. .: **. :
MMAR_0783|M.marinum_M WKFVLVVLGVWFGAAQVGLSLFYWWFHTI---DKTLPVFVVLVYLVLCAV
MUL_1419|M.ulcerans_Agy99 WKFVLVVLGVWFGAAQVGLSLFYWWFHTI---DKTLPVFVVLVYLVLCAV
Mb0470|M.bovis_AF2122/97 WKFVLVVLGVWIGAGEVGLSLFYWWYHTL---DKTAAVFVVLVYVVACTV
Rv0461|M.tuberculosis_H37Rv WKFVLVVLGVWIGAGEVGLSLFYWWYHTL---DKTAAVFVVLVYVVACTV
MAV_4688|M.avium_104 WKLVLVLLAVWAASAVVGLGLFSWWYHSI---DKTPALFAVLVFVVVCVV
Mflv_0120|M.gilvum_PYR-GCK WRFVLVTAGMWIAAAAAGAGLYYWWFHGA---DKSLPVFAVLVFVVACTV
Mvan_0793|M.vanbaalenii_PYR-1 WRFVLVVAGMWIVAAAAGAGLYYWWYHSP---DKAVPVFAVLVFVVACIV
TH_2920|M.thermoresistible__bu WVFALVVAGVWTVAAAGGLGLYHWWYVSL---DKTPTVATLLIYLMVSTV
MAB_4128c|M.abscessus_ATCC_199 WVLLLILVGTWIPAAAVGLLLYQNWFATLLPHDRIVAVEYVLAWVFACVL
* : *: . * :. * *: *: *: .: :* ::. . :
MMAR_0783|M.marinum_M ASLLLSMVQGRPLITALSLALMSAPFASVAAAAPLYGSYFCEHASRCLAG
MUL_1419|M.ulcerans_Agy99 ASLLLSMVQGRPLITALSLALMSAPFASVAAAAPLYGSYFCEHASRCLAG
Mb0470|M.bovis_AF2122/97 GGLILALVPGRPLITALSLGVMSGPFASVAAAAPLYGYYYCERMSHCLVG
Rv0461|M.tuberculosis_H37Rv GGLILALVPGRPLITALSLGVMSGPFASVAAAAPLYGYYYCERMSHCLVG
MAV_4688|M.avium_104 GGVMLAMVEARPLISALAVAVLSGPFAALAAAAPLYGYYYCARVGHCLVG
Mflv_0120|M.gilvum_PYR-GCK GSLLTAMVQERPGLAAMSLALMSAPLAAALGAGGLYASYVFGWIST----
Mvan_0793|M.vanbaalenii_PYR-1 GSLLTAMVQDRPGVAATAIAMMSAPLAAAVGAAVLYGAYVFEWIAR----
TH_2920|M.thermoresistible__bu AGMLLAMVQTKPVVSGLAIALMSAPFASTMAAAALHGAYYFNWLNP----
MAB_4128c|M.abscessus_ATCC_199 IALLLAMFEPRPLIVALAFAVLTAPAISVAAAGVKYGYVACTAPSVPGVK
.:: ::. :* : . :..:::.* : .*. :.
MMAR_0783|M.marinum_M VIPY--------------
MUL_1419|M.ulcerans_Agy99 VIPY--------------
Mb0470|M.bovis_AF2122/97 VIPY--------------
Rv0461|M.tuberculosis_H37Rv VIPY--------------
MAV_4688|M.avium_104 VVPY--------------
Mflv_0120|M.gilvum_PYR-GCK ------------------
Mvan_0793|M.vanbaalenii_PYR-1 ------------------
TH_2920|M.thermoresistible__bu ------------------
MAB_4128c|M.abscessus_ATCC_199 SLCPEPLRSWGIQYRDRA