For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADDKDASSPGTEAFVPDFSDDDDAADTGTQSWAPDFDDDAEDDDTEAAEPAAERGPEPEPEPEPAGFAGP VPPVAVPGRYFYVKWWKLVLVLLAVWAASAVVGLGLFSWWYHSIDKTPALFAVLVFVVVCVVGGVMLAMV EARPLISALAVAVLSGPFAALAAAAPLYGYYYCARVGHCLVGVVPY*
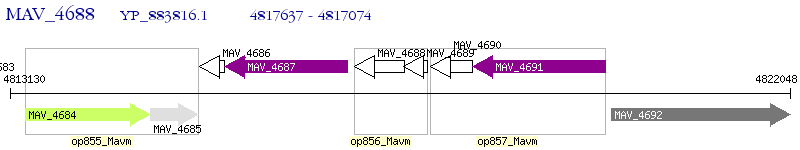
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4688 | - | - | 100% (187) | hypothetical protein MAV_4688 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0470 | - | 2e-47 | 52.02% (173) | hypothetical protein Mb0470 |
| M. gilvum PYR-GCK | Mflv_0120 | - | 6e-23 | 42.48% (113) | hypothetical protein Mflv_0120 |
| M. tuberculosis H37Rv | Rv0461 | - | 2e-47 | 52.02% (173) | hypothetical protein Rv0461 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4128c | - | 3e-18 | 36.57% (175) | hypothetical protein MAB_4128c |
| M. marinum M | MMAR_0783 | - | 9e-47 | 46.38% (207) | transmembrane protein |
| M. smegmatis MC2 155 | MSMEG_0766 | purT | 9e-05 | 38.71% (62) | phosphoribosylglycinamide formyltransferase 2 |
| M. thermoresistible (build 8) | TH_2920 | - | 1e-28 | 38.38% (185) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1419 | - | 7e-46 | 48.40% (188) | transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_0793 | - | 2e-25 | 50.00% (102) | hypothetical protein Mvan_0793 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0470|M.bovis_AF2122/97 --------------------------------------------------
Rv0461|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0783|M.marinum_M --------------------------------------------------
MUL_1419|M.ulcerans_Agy99 --------------------------------------------------
MAV_4688|M.avium_104 --------------------------------------------------
Mflv_0120|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0793|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2920|M.thermoresistible__bu --------------------------------------------------
MAB_4128c|M.abscessus_ATCC_199 ---------MSDKNAENTVEKTDAEADGAAAELVDDADQPSVVEGDLADP
MSMEG_0766|M.smegmatis_MC2_155 MSESIEGALTDDAAADESAGAGPDDTDVTDDSVADTAPEDTAPDPDDTEP
Mb0470|M.bovis_AF2122/97 ------MPDFDTGAHSQRFLS----------LAGQQ---DRAGKSWPG--
Rv0461|M.tuberculosis_H37Rv ------MPDFDTGAHSQRFLS----------LAGQQ---DRAGKSWPG--
MMAR_0783|M.marinum_M ----MAKDDLDSNPGTEPFVPEFDTGERSVPFVPDF---DTDSKPFAGLA
MUL_1419|M.ulcerans_Agy99 ----MAKDDLDSNPGTEP-------------FVPDF---DMDSKPFAGLA
MAV_4688|M.avium_104 ----MADDKDASSPGTEAFVP---------DFSDDDDAADTGTQSWAP--
Mflv_0120|M.gilvum_PYR-GCK -------------------------------MS-EQTILEDPS-------
Mvan_0793|M.vanbaalenii_PYR-1 -------------------------------MSEDQIALDDPA-------
TH_2920|M.thermoresistible__bu --VPDRRSETLPGEPTGQYTP------VFDTGSFTAVQPDDPARVWEA-D
MAB_4128c|M.abscessus_ATCC_199 LAVPIDDGKARDGLDTATFTAFSAFKDDEDDLVPTG-RIDTDALPLTEAF
MSMEG_0766|M.smegmatis_MC2_155 GDRAAADQETDTDPEPQPVVGPDAADHRPTVMLLGGGALSRELALAFQNL
.
Mb0470|M.bovis_AF2122/97 --------------STPKPQEDPVGVAP-SASVEVLG---------SEPA
Rv0461|M.tuberculosis_H37Rv --------------STPKPQEDPVGVAP-SASVEVLG---------SEPA
MMAR_0783|M.marinum_M ----------AVVTESKKSSVELVDLSEVSAPLTTLDGDSLDGDSPAEGT
MUL_1419|M.ulcerans_Agy99 ----------AVVTESKKSSVELVDLSEVSAPLTTLDGDSLDGDSPAEGT
MAV_4688|M.avium_104 ----------DFDDDAEDDDTEAAEPAAERGPEPEPEPE-------PAGF
Mflv_0120|M.gilvum_PYR-GCK -------------------------------PSNELP-------------
Mvan_0793|M.vanbaalenii_PYR-1 -------------------------------YADELP-------------
TH_2920|M.thermoresistible__bu ----------PVKPLAAEFEPATDSPQTGDVPIGELPS---------EPI
MAB_4128c|M.abscessus_ATCC_199 ----------DVGSDRTGAIPAAVGRSRAEAMTPDPYTG-----AVPVLG
MSMEG_0766|M.smegmatis_MC2_155 GGVVVAVDRYDAPAHGVADRTAVVDLNDADELSAVIAREKPQYVVAAESG
Mb0470|M.bovis_AF2122/97 ATLAHSVTVPG---RYTYLKWWKFVLVVLG----VWIGAGEVGLSLFYWW
Rv0461|M.tuberculosis_H37Rv ATLAHSVTVPG---RYTYLKWWKFVLVVLG----VWIGAGEVGLSLFYWW
MMAR_0783|M.marinum_M ATPAPSVTVPS---RYVYLKWWKFVLVVLG----VWFGAAQVGLSLFYWW
MUL_1419|M.ulcerans_Agy99 ATPAPSVTVPS---RYVYLKWWKFVLVVLG----VWFGAAQVGLSLFYWW
MAV_4688|M.avium_104 AGPVPPVAVPG---RYFYVKWWKLVLVLLA----VWAASAVVGLGLFSWW
Mflv_0120|M.gilvum_PYR-GCK PPHAQPIVVPS---AEQSVSRWRFVLVTAG----MWIAAAAAGAGLYYWW
Mvan_0793|M.vanbaalenii_PYR-1 RPPAQPVVVPA---AAQSVGRWRFVLVVAG----MWIVAAAAGAGLYYWW
TH_2920|M.thermoresistible__bu PAPARPMVVPG---SYQFLKRWVFALVVAG----VWTVAAAGGLGLYHWW
MAB_4128c|M.abscessus_ATCC_199 DEPAKPVRVPAER-EPALLKRWVLLLILVG----TWIPAAAVGLLLYQNW
MSMEG_0766|M.smegmatis_MC2_155 VIAADALIAAAERGDSEVLPTPRSIRLSQNREGLRRLASDELGLPTVPFW
. .: ... : : : * *
Mb0470|M.bovis_AF2122/97 YHTL---DKTAAVFVVLVYVVACTVGGL---ILALVPGRPLITALSLGVM
Rv0461|M.tuberculosis_H37Rv YHTL---DKTAAVFVVLVYVVACTVGGL---ILALVPGRPLITALSLGVM
MMAR_0783|M.marinum_M FHTI---DKTLPVFVVLVYLVLCAVASL---LLSMVQGRPLITALSLALM
MUL_1419|M.ulcerans_Agy99 FHTI---DKTLPVFVVLVYLVLCAVASL---LLSMVQGRPLITALSLALM
MAV_4688|M.avium_104 YHSI---DKTPALFAVLVFVVVCVVGGV---MLAMVEARPLISALAVAVL
Mflv_0120|M.gilvum_PYR-GCK FHGA---DKSLPVFAVLVFVVACTVGSL---LTAMVQERPGLAAMSLALM
Mvan_0793|M.vanbaalenii_PYR-1 YHSP---DKAVPVFAVLVFVVACIVGSL---LTAMVQDRPGVAATAIAMM
TH_2920|M.thermoresistible__bu YVSL---DKTPTVATLLIYLMVSTVAGM---LLAMVQTKPVVSGLAIALM
MAB_4128c|M.abscessus_ATCC_199 FATLLPHDRIVAVEYVLAWVFACVLIAL---LLAMFEPRPLIVALAFAVL
MSMEG_0766|M.smegmatis_MC2_155 FAGSVEELTAVAEHAGYPLVVKPVTGSLRDGQSVLLRPDDIEPAWKRATT
: . :. .: :. . .
Mb0470|M.bovis_AF2122/97 SGPFAS--VAAAAPLYGYYYCERMSHCLVGVIPY----------------
Rv0461|M.tuberculosis_H37Rv SGPFAS--VAAAAPLYGYYYCERMSHCLVGVIPY----------------
MMAR_0783|M.marinum_M SAPFAS--VAAAAPLYGSYFCEHASRCLAGVIPY----------------
MUL_1419|M.ulcerans_Agy99 SAPFAS--VAAAAPLYGSYFCEHASRCLAGVIPY----------------
MAV_4688|M.avium_104 SGPFAA--LAAAAPLYGYYYCARVGHCLVGVVPY----------------
Mflv_0120|M.gilvum_PYR-GCK SAPLAA--ALGAGGLYASYVFGWIST------------------------
Mvan_0793|M.vanbaalenii_PYR-1 SAPLAA--AVGAAVLYGAYVFEWIAR------------------------
TH_2920|M.thermoresistible__bu SAPFAS--TMAAAALHGAYYFNWLNP------------------------
MAB_4128c|M.abscessus_ATCC_199 TAPAIS--VAAAGVKYGYVACTAPSVPGVKSLCPEPLRSWGIQYRDRA--
MSMEG_0766|M.smegmatis_MC2_155 AGHFAHSRVLAESVVEVDFEVTMLTIRTTGPTGPGVQFCEPIGHRRSDNG
:. . .
Mb0470|M.bovis_AF2122/97 --------------------------------------------------
Rv0461|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0783|M.marinum_M --------------------------------------------------
MUL_1419|M.ulcerans_Agy99 --------------------------------------------------
MAV_4688|M.avium_104 --------------------------------------------------
Mflv_0120|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0793|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2920|M.thermoresistible__bu --------------------------------------------------
MAB_4128c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0766|M.smegmatis_MC2_155 VLESWQPQQMPPAALDSAKSIAARIVNSLGGRGVFGVELLVQGEDVYFSD
Mb0470|M.bovis_AF2122/97 --------------------------------------------------
Rv0461|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0783|M.marinum_M --------------------------------------------------
MUL_1419|M.ulcerans_Agy99 --------------------------------------------------
MAV_4688|M.avium_104 --------------------------------------------------
Mflv_0120|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0793|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2920|M.thermoresistible__bu --------------------------------------------------
MAB_4128c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0766|M.smegmatis_MC2_155 VRVRPHDSGLVTLRSQRLSEFELHARAILGLACDTIMISPGAAEVTYAGA
Mb0470|M.bovis_AF2122/97 --------------------------------------------------
Rv0461|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0783|M.marinum_M --------------------------------------------------
MUL_1419|M.ulcerans_Agy99 --------------------------------------------------
MAV_4688|M.avium_104 --------------------------------------------------
Mflv_0120|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0793|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2920|M.thermoresistible__bu --------------------------------------------------
MAB_4128c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0766|M.smegmatis_MC2_155 EAAPSVDVTAVLGEALATSESDVRLFGRAEESDDGRRLGVALATAPDVIV
Mb0470|M.bovis_AF2122/97 ----------------
Rv0461|M.tuberculosis_H37Rv ----------------
MMAR_0783|M.marinum_M ----------------
MUL_1419|M.ulcerans_Agy99 ----------------
MAV_4688|M.avium_104 ----------------
Mflv_0120|M.gilvum_PYR-GCK ----------------
Mvan_0793|M.vanbaalenii_PYR-1 ----------------
TH_2920|M.thermoresistible__bu ----------------
MAB_4128c|M.abscessus_ATCC_199 ----------------
MSMEG_0766|M.smegmatis_MC2_155 ARDRARRVGTALRRLW