For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GGDAHPQEVEFLRDARRIAMAHRGFTSFRFPMNSMGAFHEAAKIGFRYIETDVRATRDGVAVIQHDRKLA PESGVAGAVDQLTWPEVSTADLGGGEPIPTLEELLVALPQMRFNIDIKADSAVEPTVEVIERLQAHRRVL IASFSERRRQRALRLMSQRVASAAGMGAFLGFMAARTAGRRASAMRMLRDSDCLQLPARFGGLPVITPAL VRSVHASRRQVHAWTIDDPAMMHALFDIGVDGIITDRADVLCDVLGARGEW*
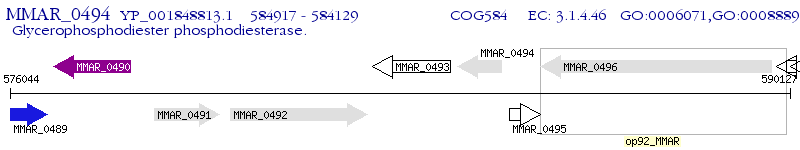
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0494 | glpQ2 | - | 100% (262) | glycerophosphoryl diester phosphodiesterase GlpQ2 |
| M. marinum M | MMAR_5394 | glpQ1 | 4e-08 | 26.77% (254) | glycerophosphoryl diester phosphodiesterase GlpQ1 |
| M. marinum M | MMAR_4162 | gdpD | 1e-05 | 26.99% (163) | glycerolphosphodiesterase GdpD |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0325c | glpQ2 | 1e-101 | 71.94% (253) | putative glycerophosphoryl diester phosphodiesterase GLPQ2 |
| M. gilvum PYR-GCK | Mflv_1140 | - | 1e-08 | 25.97% (258) | glycerophosphodiester phosphodiesterase |
| M. tuberculosis H37Rv | Rv0317c | glpQ2 | 1e-101 | 71.94% (253) | glycerophosphoryl diester phosphodiesterase |
| M. leprae Br4923 | MLBr_00074 | glpQ | 1e-08 | 29.39% (245) | putative glycerophosphoryl diester phosphodiesterase |
| M. abscessus ATCC 19977 | MAB_0123 | - | 4e-09 | 26.56% (256) | putative glycerophosphoryl diester phosphodiesterase |
| M. avium 104 | MAV_0576 | - | 2e-08 | 26.39% (288) | glycerophosphoryl diester phosphodiesterase family protein |
| M. smegmatis MC2 155 | MSMEG_6423 | - | 4e-09 | 27.91% (258) | glycerophosphoryl diester phosphodiesterase family protein |
| M. thermoresistible (build 8) | TH_0749 | glpQ1 | 6e-07 | 34.38% (96) | PROBABLE GLYCEROPHOSPHORYL DIESTER PHOSPHODIESTERASE GLPQ1 |
| M. ulcerans Agy99 | MUL_1157 | glpQ2 | 1e-139 | 98.02% (253) | glycerophosphoryl diester phosphodiesterase GlpQ2 |
| M. vanbaalenii PYR-1 | Mvan_5667 | - | 3e-08 | 26.15% (260) | glycerophosphoryl diester phosphodiesterase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1140|M.gilvum_PYR-GCK ---------------------MSSGATLGG--HPFVVAHRGASADR-PEH
Mvan_5667|M.vanbaalenii_PYR-1 ---------------------MSSGETFGG--HPFVVAHRGASADR-PEH
MSMEG_6423|M.smegmatis_MC2_155 ------MTEAD---------AGADGQAPTGPTHPFVVAHRGASADR-PEH
MAB_0123|M.abscessus_ATCC_1997 ------MTESSEPGEDEAHARGADGIDGRGERHPFVVAHRGASADR-PEH
MLBr_00074|M.leprae_Br4923 --------------------MMLADEVFAG--HPFVVAHRGASAVL-PEH
TH_0749|M.thermoresistible__bu --------------------METGGEALGR--HPYVVAHRGASADR-PEH
MMAR_0494|M.marinum_M ----------------MGGDAHPQEVEFLRDARRIAMAHRGFTSFRFPMN
MUL_1157|M.ulcerans_Agy99 -------------------------MEFLRDARRIAIAHRGFTSFRFPMN
Mb0325c|M.bovis_AF2122/97 -------------------------MEFLRHGGRIAMAHRGFTSFRLPMN
Rv0317c|M.tuberculosis_H37Rv -------------------------MEFLRHGGRIAMAHRGFTSFRLPMN
MAV_0576|M.avium_104 MSSLGSRWAVAAAVALLMAVLLSAAPVYGQPATFDLQAHRGGRGET-TEE
**** . . .
Mflv_1140|M.gilvum_PYR-GCK TLAAYQLALEEGADAVECDVRLTRDGHLVCVHDRRLDRTSSGTG------
Mvan_5667|M.vanbaalenii_PYR-1 TLAAYELALGEGADAVECDVRLTRDGHLVCVHDRRLDRTSTGSG------
MSMEG_6423|M.smegmatis_MC2_155 TLAAYDLALQEGADGLECDVRLTRDGHLVCVHDRKVDRTSTGTG------
MAB_0123|M.abscessus_ATCC_1997 TLAAYELALEEGADGVECDVRLTRDGQLVCVHDRRVDRTSDGTG------
MLBr_00074|M.leprae_Br4923 TLAAYELALKEGADGVECDVRLTRDGHMVCVHDRRLDRTSTGVG------
TH_0749|M.thermoresistible__bu TLAAYELALEEGADGLECDVRLTRDGQLVCVHDRRVDRTSTGSG------
MMAR_0494|M.marinum_M SMGAFHEAAKIGFRYIETDVRATRDGVAVIQHDRKLAPESGVAG------
MUL_1157|M.ulcerans_Agy99 SMGAFHEAAKIGFRYIETDVRATRDGVAVIQHDRKLTPESGVAG------
Mb0325c|M.bovis_AF2122/97 SMGAFQEAAKLGFRYIETDVRATRDGVAVILHDRRLAPGVGLSG------
Rv0317c|M.tuberculosis_H37Rv SMGAFQEAAKLGFRYIETDVRATRDGVAVILHDRRLAPGVGLSG------
MAV_0576|M.avium_104 SLRAFAKAIQTGVSTLELDIVVTRDGQPLVWHDPTIQADKCSDTGPAFPG
:: *: * * :* *: **** : ** :
Mflv_1140|M.gilvum_PYR-GCK ---------LVSDMTLAQLRELDFGAWH--PSRRDDVSGGDTGLLTLDDL
Mvan_5667|M.vanbaalenii_PYR-1 ---------LVSEMTLAQLRDFDFGSWH--PSWRADGSHGDTGLLTLDDL
MSMEG_6423|M.smegmatis_MC2_155 ---------LVSEMTLAELRKLNFGSWH--PSWRADSSGDDTGVLTLDEL
MAB_0123|M.abscessus_ATCC_1997 ---------LVSEMTLSQLRALDFGSWH--PGGAEAETG--TGLLTLEEL
MLBr_00074|M.leprae_Br4923 ---------LVSTMTLAQLRALEYGAWH--NSWRPDGTHGDTGLLTLDAL
TH_0749|M.thermoresistible__bu ---------VISTMTLDELRRLDFGAWH--DSWQP--VYGTFGLLTLEEL
MMAR_0494|M.marinum_M ---------AVDQLTWPEVSTADLGGGEPIPTLEELLVALPQMRFNIDIK
MUL_1157|M.ulcerans_Agy99 ---------AVDQLTWPEVSIADLGGGEPIPTLEELLVALPQMRFNIDIK
Mb0325c|M.bovis_AF2122/97 ---------AVDRLDWRDVRKAQLGAGQSIPTLEDLLTALPDMRVNIDIK
Rv0317c|M.tuberculosis_H37Rv ---------AVDRLDWRDVRKAQLGAGQSIPTLEDLLTALPDMRVNIDIK
MAV_0576|M.avium_104 DPGYPYVGKLVHQLTVAQIHTLDCGHRL--KEFPDAEVVRGNKIATLPEV
: : :: : * .:
Mflv_1140|M.gilvum_PYR-GCK VSLVMDWNRPVKMFIETKHPVRYGGLVESKVLALLHRYGIASPASADMAR
Mvan_5667|M.vanbaalenii_PYR-1 VRLVLDWNRPVKMFIETKHPVRYGALVESKVLALLHRYGIASPASADLSR
MSMEG_6423|M.smegmatis_MC2_155 VRMALDWNRPVKLFIETKHPVRYGALVENKLLALLHRYGIAAPASADMAR
MAB_0123|M.abscessus_ATCC_1997 VSLVLDWNRPVKLFIETKHPVRYGSLVENKVLALLHRFGIAAPASADMSR
MLBr_00074|M.leprae_Br4923 VSLVLDWHRPVKIFVETKHPVRYGALVESKLLALLHRFGIAAPASADRSR
TH_0749|M.thermoresistible__bu IGLVLGWKRPVKLFIETKHPVRYGGLVESKVLAMLHRFGIASPPSARLSR
MMAR_0494|M.marinum_M ADSAVEPTVEVIERLQAHRRVLIASFSERRRQRALRLMSQRVASAAGMG-
MUL_1157|M.ulcerans_Agy99 ADSAVEPTVEVIERLQAHRRVLIASFSERRRQRALRLMSQRVASAAGMG-
Mb0325c|M.bovis_AF2122/97 AASAIEPTVNVIERCNAHNRVLIGSFSERRRRRALRLLTKRVASSAGTG-
Rv0317c|M.tuberculosis_H37Rv AASAIEPTVNVIERCNAHNRVLIGSFSERRRRRALRLLTKRVASSAGTG-
MAV_0576|M.avium_104 FAVADSHRADAIRYNIETKVEPDDTGASAPPQEFVDVILAAVRSADKLAR
. . . . : .: .
Mflv_1140|M.gilvum_PYR-GCK AVVMSFSAGAVWRIRRAAPMLPTVLLGETSRYLGGGAATTVGATAVGPSI
Mvan_5667|M.vanbaalenii_PYR-1 AVVMSFSAAAVWRIRRAAPLLPTVLLGETSRYLGGSAATTVGATAVGPSI
MSMEG_6423|M.smegmatis_MC2_155 AVVMSFSAAAVWRIRRAAPMLPTVLLGETSRYLGGSAATTVGATAVGPSI
MAB_0123|M.abscessus_ATCC_1997 AVVMSFSAAAVWRIRRAAPMLPTVLLGETSRYLGGSAATTVGATAVGPSI
MLBr_00074|M.leprae_Br4923 AVVMSFSAAAVWRVRRAAPLLPTVLLGRAGRYLTSSAATAVGATAVGPSL
TH_0749|M.thermoresistible__bu AVIMX-------GRRRSTTWRPA---------------------------
MMAR_0494|M.marinum_M AFLGFMAARTAGRRASAMRMLRDSDCLQLPARFGG---------------
MUL_1157|M.ulcerans_Agy99 VFLGFMAARTAGRRASAMRMLRDSDRLQLPARFGG---------------
Mb0325c|M.bovis_AF2122/97 ALLAWLTARPLGSRAYAWRMMRDIDCVQLPSRLGG---------------
Rv0317c|M.tuberculosis_H37Rv ALLAWLTARPLGSRAYAWRMMRDIDCVQLPSRLGG---------------
MAV_0576|M.avium_104 VEIQSFDWRTLPLVRRAEPSIPLVALYDERAAGDPVLGARAAGADIVSPD
. : :
Mflv_1140|M.gilvum_PYR-GCK ATLREHPELVDRAAAQGRALYCWTVDHYEDVQYCADLGVAWIATNHPGRT
Mvan_5667|M.vanbaalenii_PYR-1 ATLREHPELVDRAAAQGRALYCWTVDHYEDVEYCRDLGVAWIATNHPGRT
MSMEG_6423|M.smegmatis_MC2_155 ATLREHPELVDRAAAQGRALYCWTVDHYEDVRFCRDVGVAWIATNHPGRT
MAB_0123|M.abscessus_ATCC_1997 ATLREHPELVDRAAAQGRATYCWTVDHYEDVEFCRSIGVAWIATNHPGRT
MLBr_00074|M.leprae_Br4923 PALKEYPKLVDRAGAQGRAVYCWNVDDYDDIDFCRDIGVAWIATRHPGRT
TH_0749|M.thermoresistible__bu --------------------------------------------------
MMAR_0494|M.marinum_M -LPVITPALVRSVHASRRQVHAWTIDDPAMMHALFDIGVDGIITDRADVL
MUL_1157|M.ulcerans_Agy99 -LPVITPALVRSVHASRRQVHAWTIDDPAMMHALFDIGVDGIITDRADVL
Mb0325c|M.bovis_AF2122/97 -VPVITPARVRGFHAAGRQVHAWTVDEPDVMHTLLDMDVDGIITDRADLL
Rv0317c|M.tuberculosis_H37Rv -VPVITPARVRGFHAAGRQVHAWTVDEPDVMHTLLDMDVDGIITDRADLL
MAV_0576|M.avium_104 YRLVTGKPYVDRAHALGLKVIPWTVDDAENMRRQIDYGVDGLITDFPTLA
Mflv_1140|M.gilvum_PYR-GCK KAWLQNGLTGAGRD---
Mvan_5667|M.vanbaalenii_PYR-1 KGWLQDGLTGAGRD---
MSMEG_6423|M.smegmatis_MC2_155 KDWLQNGLTGAGRD---
MAB_0123|M.abscessus_ATCC_1997 KSWLQRGLTGTTL----
MLBr_00074|M.leprae_Br4923 KAWLQN--NGRTE----
TH_0749|M.thermoresistible__bu -----------------
MMAR_0494|M.marinum_M CDVLGARGEW-------
MUL_1157|M.ulcerans_Agy99 CDVLGARGEW-------
Mb0325c|M.bovis_AF2122/97 RDVLIARGEWDGA----
Rv0317c|M.tuberculosis_H37Rv RDVLIARGEWDGA----
MAV_0576|M.avium_104 RGVLAELGVPLPPAVPG