For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSSLGSRWAVAAAVALLMAVLLSAAPVYGQPATFDLQAHRGGRGETTEESLRAFAKAIQTGVSTLELDIV VTRDGQPLVWHDPTIQADKCSDTGPAFPGDPGYPYVGKLVHQLTVAQIHTLDCGHRLKEFPDAEVVRGNK IATLPEVFAVADSHRADAIRYNIETKVEPDDTGASAPPQEFVDVILAAVRSADKLARVEIQSFDWRTLPL VRRAEPSIPLVALYDERAAGDPVLGARAAGADIVSPDYRLVTGKPYVDRAHALGLKVIPWTVDDAENMRR QIDYGVDGLITDFPTLARGVLAELGVPLPPAVPG
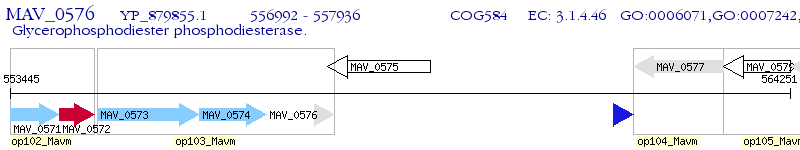
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0576 | - | - | 100% (314) | glycerophosphoryl diester phosphodiesterase family protein |
| M. avium 104 | MAV_0186 | - | 3e-12 | 28.90% (263) | glycerophosphoryl diester phosphodiesterase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3872c | glpQ1 | 4e-11 | 28.52% (263) | glycerophosphoryl diester phosphodiesterase |
| M. gilvum PYR-GCK | Mflv_1140 | - | 1e-14 | 30.22% (278) | glycerophosphodiester phosphodiesterase |
| M. tuberculosis H37Rv | Rv3842c | glpQ1 | 4e-11 | 28.52% (263) | glycerophosphoryl diester phosphodiesterase |
| M. leprae Br4923 | MLBr_00325 | - | 8e-14 | 59.38% (64) | hypothetical protein MLBr_00325 |
| M. abscessus ATCC 19977 | MAB_0573 | - | 1e-101 | 56.88% (327) | putative glycerophosphoryl diester phosphodiesterase precursor |
| M. marinum M | MMAR_5394 | glpQ1 | 5e-13 | 29.33% (283) | glycerophosphoryl diester phosphodiesterase GlpQ1 |
| M. smegmatis MC2 155 | MSMEG_6423 | - | 3e-15 | 30.68% (264) | glycerophosphoryl diester phosphodiesterase family protein |
| M. thermoresistible (build 8) | TH_0749 | glpQ1 | 7e-05 | 42.22% (45) | PROBABLE GLYCEROPHOSPHORYL DIESTER PHOSPHODIESTERASE |
| M. ulcerans Agy99 | MUL_5015 | glpQ1 | 3e-12 | 29.27% (287) | glycerophosphoryl diester phosphodiesterase GlpQ1 |
| M. vanbaalenii PYR-1 | Mvan_0998 | - | 1e-108 | 59.76% (333) | glycerophosphoryl diester phosphodiesterase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3872c|M.bovis_AF2122/97 ---------------MTWADEVLAG-----------------HPFVVAHR
Rv3842c|M.tuberculosis_H37Rv ---------------MTWADEVLAG-----------------HPFVVAHR
MMAR_5394|M.marinum_M -------------MGMTWADEVIAG-----------------HPFVVAHR
MUL_5015|M.ulcerans_Agy99 ---------------MTWADEVIAG-----------------HPFVVAHR
Mflv_1140|M.gilvum_PYR-GCK ----------------MSSGATLGG-----------------HPFVVAHR
MSMEG_6423|M.smegmatis_MC2_155 ----------MTEADAGADGQAPTGPT---------------HPFVVAHR
TH_0749|M.thermoresistible__bu ---------------METGGEALGR-----------------HPYVVAHR
MAB_0573|M.abscessus_ATCC_1997 -----------MKRIAAVLAVTLALVS--------CGDKDTKSFDLQAHR
Mvan_0998|M.vanbaalenii_PYR-1 MPSNTAAPPQASRRRRLLLAAVTAMVVGSVPTVSFPAAAQVPEFDLQAHR
MAV_0576|M.avium_104 -MSSLGSRWAVAAAVALLMAVLLSAAP---------VYGQPATFDLQAHR
MLBr_00325|M.leprae_Br4923 --------------------------------------------------
Mb3872c|M.bovis_AF2122/97 GASAARPEHTLAAYDLALKEGADGVECDVRLTRDGHLVCVHDRRLDRTST
Rv3842c|M.tuberculosis_H37Rv GASAARPEHTLAAYDLALKEGADGVECDVRLTRDGHLVCVHDRRLDRTST
MMAR_5394|M.marinum_M GASGARPEHTLAAYDLALKQGADGVECDVRLTRDGHLVCVHDRRLDRTST
MUL_5015|M.ulcerans_Agy99 GASGARPEHTLAAYDLALKQGADGVECDVRLTRDGHLVCVHDRRLDRTST
Mflv_1140|M.gilvum_PYR-GCK GASADRPEHTLAAYQLALEEGADAVECDVRLTRDGHLVCVHDRRLDRTSS
MSMEG_6423|M.smegmatis_MC2_155 GASADRPEHTLAAYDLALQEGADGLECDVRLTRDGHLVCVHDRKVDRTST
TH_0749|M.thermoresistible__bu GASADRPEHTLAAYELALEEGADGLECDVRLTRDGQLVCVHDRRVDRTST
MAB_0573|M.abscessus_ATCC_1997 GGRGETTEESLRAFAKALELGVSTLELDIVISKDGKPMVWHDPVIDPAKC
Mvan_0998|M.vanbaalenii_PYR-1 GGRGETTEESLRAFAKSLELGVSTLELDVVLTKDGQPLVWHDPVLQPEKC
MAV_0576|M.avium_104 GGRGETTEESLRAFAKAIQTGVSTLELDIVVTRDGQPLVWHDPTIQADKC
MLBr_00325|M.leprae_Br4923 --------------------------------------------------
Mb3872c|M.bovis_AF2122/97 GAG---------------LVSTMTLAQLRELEYGAWHDSWRPDGSHGDTS
Rv3842c|M.tuberculosis_H37Rv GAG---------------LVSTMTLAQLRELEYGAWHDSWRPDGSHGDTS
MMAR_5394|M.marinum_M GAG---------------LVSTMTLDQLRELEYGAWHDSWRPDGAHGDTS
MUL_5015|M.ulcerans_Agy99 GAG---------------LVSTMTLDQLRELEYGAWHDSRRPDGAHGDTS
Mflv_1140|M.gilvum_PYR-GCK GTG---------------LVSDMTLAQLRELDFGAWHPSRRDDVSGGDTG
MSMEG_6423|M.smegmatis_MC2_155 GTG---------------LVSEMTLAELRKLNFGSWHPSWRADSSGDDTG
TH_0749|M.thermoresistible__bu GSG---------------VISTMTLDELRRLDFGAWHDSWQP--VYGTFG
MAB_0573|M.abscessus_ATCC_1997 SDTGPVRPGDPQFPYVGKLVKDLTSGQLHTLDCGKLLTDFPRAEVVKNDK
Mvan_0998|M.vanbaalenii_PYR-1 TDTGPAFAADAQYPYVGKLVHDLTLAQIRTLDCGKLLDDFPDAEVVTANK
MAV_0576|M.avium_104 SDTGPAFPGDPGYPYVGKLVHQLTVAQIHTLDCGHRLKEFPDAEVVRGNK
MLBr_00325|M.leprae_Br4923 --------------------------------------------MLRGNK
Mb3872c|M.bovis_AF2122/97 LLTLDALVSLVLDWHRP-VKIFVETKHPVRYGSLVENKLLALLHRFGIAA
Rv3842c|M.tuberculosis_H37Rv LLTLDALVSLVLDWHRP-VKIFVETKHPVRYGSLVENKLLALLHRFGIAA
MMAR_5394|M.marinum_M LLTLDALISLVLDWKRP-VKVFIETKHPVRYGSLVESKVLALLQRFGIAQ
MUL_5015|M.ulcerans_Agy99 LLTLDALISLVLDWKRP-VKVFIETKHPVRYGSLVESKVLALLQRFGIAQ
Mflv_1140|M.gilvum_PYR-GCK LLTLDDLVSLVMDWNRP-VKMFIETKHPVRYGGLVESKVLALLHRYGIAS
MSMEG_6423|M.smegmatis_MC2_155 VLTLDELVRMALDWNRP-VKLFIETKHPVRYGALVENKLLALLHRYGIAA
TH_0749|M.thermoresistible__bu LLTLEELIGLVLGWKRP-VKLFIETKHPVRYGGLVESKVLAMLHRFGIAS
MAB_0573|M.abscessus_ATCC_1997 IAQLSDVFALADSYHAD-VRFNIETKVEAAE-PQKSAEPQRFVDIILETI
Mvan_0998|M.vanbaalenii_PYR-1 IATLPEVFGLADSYAAD-VRFNIETKIEADK-PETSAGPREIVDAVLAAA
MAV_0576|M.avium_104 IATLPEVFAVADSHRADAIRYNIETKVEPDD-TGASAPPQEFVDVILAAV
MLBr_00325|M.leprae_Br4923 IVVLPEVFALTDYYLAD-VRFNIETKVAADR-PAVSVFSQEFVDVVLAVV
: * :. :. :: :*** . ::. .
Mb3872c|M.bovis_AF2122/97 PASADRSRAVVMSFSAAAVWRIRRAAPLLPTVLLGKTPRYLTSS------
Rv3842c|M.tuberculosis_H37Rv PASADRSRAVVMSFSAAAVWRIRRAAPLLPTVLLGKTPRYLTSS------
MMAR_5394|M.marinum_M PASADRSRAVVMSFSAAAVWRIRRAAPLLPTVLLGKTARYLASG------
MUL_5015|M.ulcerans_Agy99 PASADRSRAAAMSFSAAAVWRIRRAAPLLPTVLLGKTARYLASG------
Mflv_1140|M.gilvum_PYR-GCK PASADMARAVVMSFSAGAVWRIRRAAPMLPTVLLGETSRYLGGG------
MSMEG_6423|M.smegmatis_MC2_155 PASADMARAVVMSFSAAAVWRIRRAAPMLPTVLLGETSRYLGGS------
TH_0749|M.thermoresistible__bu PPSARLSRAVIMX-------GRRRSTTWRPA-------------------
MAB_0573|M.abscessus_ATCC_1997 RAAGKLDKVEIQSFDWRTLPMTKRAEPGIPLVALWDETTWYPGSPWLGGV
Mvan_0998|M.vanbaalenii_PYR-1 RSAGKLDRVEIQSFDWRTLPLVRHLEPSIPLVALWDETTWVPGSPWLAGI
MAV_0576|M.avium_104 RSADKLARVEIQSFDWRTLPLVRRAEPSIPLVALYDER------------
MLBr_00325|M.leprae_Br4923 RSVGKVDRVASP-----------------------GER------------
. :.
Mb3872c|M.bovis_AF2122/97 -----AATAVGATAVGPSLPALK-----------------EYPQLVDRSA
Rv3842c|M.tuberculosis_H37Rv -----AATAVGATAVGPSLPALK-----------------EYPQLVDRSA
MMAR_5394|M.marinum_M -----AASAVGATAVGPSLAVLR-----------------EYPQLVDRAA
MUL_5015|M.ulcerans_Agy99 -----AASAVGATAVGPSLAVLR-----------------EYPQLVDRAA
Mflv_1140|M.gilvum_PYR-GCK -----AATTVGATAVGPSIATLR-----------------EHPELVDRAA
MSMEG_6423|M.smegmatis_MC2_155 -----AATTVGATAVGPSIATLR-----------------EHPELVDRAA
TH_0749|M.thermoresistible__bu --------------------------------------------------
MAB_0573|M.abscessus_ATCC_1997 DPAIVSDPIDGAKQVGATILSPGYTVPYGGKVGDPGFTLVADKKFIDKAH
Mvan_0998|M.vanbaalenii_PYR-1 DPAVVGDPLTGATMVGAGIVSPGYSIPYGLTPDDPGFALVADRPFIERAH
MAV_0576|M.avium_104 ---AAGDPVLGARAAGADIVSP-------------DYRLVTGKPYVDRAH
MLBr_00325|M.leprae_Br4923 -------PADGAS-------------------------------------
Mb3872c|M.bovis_AF2122/97 AQGRAVYCWNVDEYEDIDFCREVGVAWIGTHHPGRTKAWLEDGR---ANG
Rv3842c|M.tuberculosis_H37Rv AQGRAVYCWNVDEYEDIDFCREVGVAWIGTHHPGRTKAWLEDGR---ANG
MMAR_5394|M.marinum_M AQGRAVYCWGVDEYEDFDFCRDVGVAWMATDHPGRAKAWLQHGRPGVSSG
MUL_5015|M.ulcerans_Agy99 AQGRAVYCWGVDEYEDIDFCRDVGVAWMASDHPGRAKAWLQHGRPGVSSG
Mflv_1140|M.gilvum_PYR-GCK AQGRALYCWTVDHYEDVQYCADLGVAWIATNHPGRTKAWLQNGL----TG
MSMEG_6423|M.smegmatis_MC2_155 AQGRALYCWTVDHYEDVRFCRDVGVAWIATNHPGRTKDWLQNGL----TG
TH_0749|M.thermoresistible__bu --------------------------------------------------
MAB_0573|M.abscessus_ATCC_1997 RLGLKVIPWTINDAETMKAQIDAGADGIISDYPTTLRKVMAESDLPLPPA
Mvan_0998|M.vanbaalenii_PYR-1 AMGLKVIPWTVNDEDTMRAQIVAGADGIITDYPALLRDVMTQLGLPLPPP
MAV_0576|M.avium_104 ALGLKVIPWTVDDAENMRRQIDYGVDGLITDFPTLARGVLAELGVPLPPA
MLBr_00325|M.leprae_Br4923 --GLAVY--TVG--------------GLMG--------------------
Mb3872c|M.bovis_AF2122/97 TTR----
Rv3842c|M.tuberculosis_H37Rv TTR----
MMAR_5394|M.marinum_M PAAPPEA
MUL_5015|M.ulcerans_Agy99 PAAPPEA
Mflv_1140|M.gilvum_PYR-GCK AGRD---
MSMEG_6423|M.smegmatis_MC2_155 AGRD---
TH_0749|M.thermoresistible__bu -------
MAB_0573|M.abscessus_ATCC_1997 YHR----
Mvan_0998|M.vanbaalenii_PYR-1 YRRH---
MAV_0576|M.avium_104 VPG----
MLBr_00325|M.leprae_Br4923 -------